Module: ShapeAnalysis ()
This module is deprecated. Please use Label Analysis instead.
This module computes the shape parameters of regions in the attached input data. A region is defined as a group of voxels which have the same data value. The module can be connected to a label image (or any other scalar field containing label data). As output a spreadsheet is computed that contains the following information for each detected region:
- Value: The label of the region.
- Volume: The volume of the region in units of the voxel size.
- CenterX, CenterY, CenterZ: The components of the center of mass. A weighting of 1 per voxel is assumed if no secondary Field data set is connected. If a Field data set is connected the densities in that field are used as mass.
- Anisotropy: Measures a region's deviation from a spherical shape:
If option Use all three eigenvalues is off, the following formula is used:
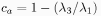
... otherwise:
- Elongation: If option Use all three eigenvalues is off, the following formula is used:
Elongated objects have small values close to 0.
... otherwise:
The closer the value to 1 the more elongated.
- Flatness: If option Use all three eigenvalues is off, the following formula is used:
Flat objects have small values close to 0.
... otherwise:
Higher values mean a greater flatness.
- EValue1, EValue2, EValue3: The eigenvalues
of the covariance matrix in descending order.
- EVector1x, EVector1y, EVector1z: Normalized eigenvector
- EVector2x, EVector2y, EVector2z: Normalized eigenvector
- EVector3x, EVector3y, EVector3z: Normalized eigenvector
- ExtentMin1, ExtentMin2, ExtentMin3, ExtentMax1, ExtentMax2, ExtentMax3: The extent of the data in the direction of the largest, medium, and smallest eigenvectors of the covariance matrix. These values are computed as dot-products of the eigenvectors with the voxel coordinates for each region. The minimum and maximum extensions are computed with respect to the center of region which is the average of the voxel positions. The sum of these values give the lengths of the minimum enclosing bounding box (see Extent1, Extent2, Extent3 below).
- Iii: Principal moments of inertia measuring the moment of inertia around the i-axis when the object is rotated around the i-axis.
- Iij | i!=j: Products of inertia measuring the moment of inertia around the i-axis if the object is rotated around the j-axis.
- Mass: If a data set is connected to the Field port this value is the sum of the intensities for each region of interest, otherwise the values stored are the number of voxels per region.
- Area: This value is the sum of voxel surfaces that are on the outside of each connected component.
- Boundary: Some components of the image volume might be intersected by the Bounding Box of the image volume. This value is the sum of voxels that are at this boundary. These voxels don't have a neighboring voxel within the volume and the component might not be fully within the image.
- Extent1, Extent2, Extent3: The lengths of the axes of the minimal enclosing bounding box.
Additionally, the spreadsheet contains a table with summary entries for the mean volume, the mean elongation, the mean flatness, and the mean orientation (with their standard deviations).
The spreadsheet can be filtered for various aspects using the module SpreadsheetFilter. Furthermore, with this module the data can be prepared for different kinds of visualizations.
Reference:
Westin CF, Peled S, Gudbjartsson H, Kikinis R, Jolesz FA. Geometrical diffusion measures for MRI from tensor basis analysis. In ISMRM '97. Vancouver Canada, 1997;1742.
Data [required]
As input a uniform scalar field with integer elements is required. This could be a LabelField or any other Scalar Field with byte, short, ushort or int32 resolution in which the values represent labels of regions. Examples are the output of ConnectedComponents or a watershed segmentation.Field [optional]
This port can be optionally connected to a gray value image. The field should be of type HxUniformScalarField3. If a data set is connected, its values multiplied with the voxel volume will be used as weight for the computation of the center of mass and the inertia tensor. If no input is provided the weight for each voxel is assumed to be 1.
Number of objects
Displays the number of regions.Mean volume
Displays the mean volume of all regions.Mean anisotropy
Displays the mean anisotropy of all regions.Mean elongation
Displays the mean elongation of all regions.Mean flatness
Displays the mean flatness of all regions.Mean orientation
Displays the mean orientation of all regions.formula
Toggles whether two or all three eigenvalues are taken into account to compute anisotropy, elongation and flatness,