It is possible to completely define the set of rules used to convert a directory in a BrainVISA database. That allows the use of BrainVISA without having to modify an existing file organization. However, the writing of such a system of rules requires a significant investment in the study of BrainVISA. This is why BrainVISA is provided with a default data organization system that can be used easily. Information about how to extend BrainVISA ontology can be found in the BrainVISA programming manual.
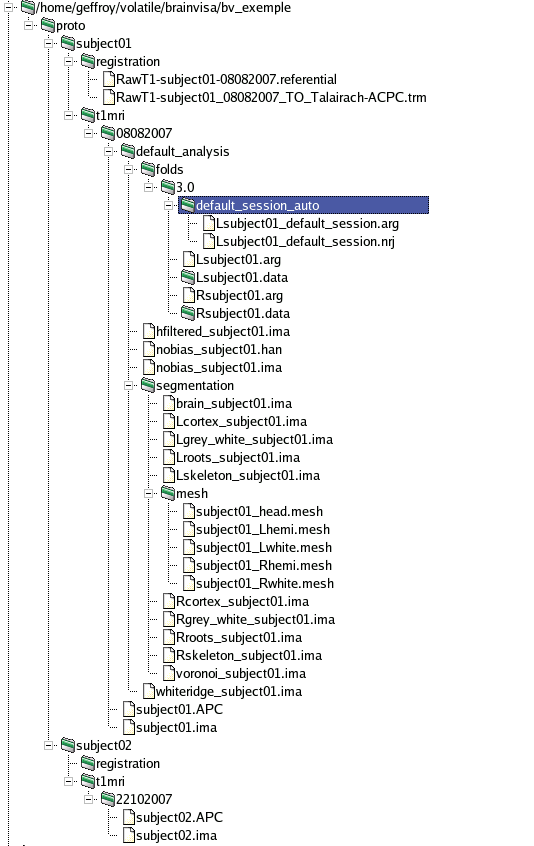
By default, data in a BrainVISA database are organized in a hierarchical structure with the following directories :
database directory
protocol: a directory for each protocol
subject: a directory for each subject
modality: for example t1mri, diffusion, pet...
acquisition: raw data. It is possible to have several acquisition for each subject.
analysis: results of an analysis, for example files generated by brainVISA during the segmentation pipeline.
T1-weighted data are stored in a directory database/protocol/subject/t1mri. Database, protocol and subject directories names are choosen by the user but t1mri is a fixed name.
t1mri directory contains :
- One or several acquisition directory, named by the user. Each contains :
- Raw data
- One or several analysis directory, named by the user, and which contains the results of an analysis on the raw data, more precisely:
- segmentation directory: contains data generated by the segmentation pipeline and a mesh directory containing meshes generated by the pipeline.
- folds directory: contains 3.0 and 3.1 directories for the different versions of sulci graphs.
Referentials and transformations for the data are stored in database/protocol/subject/registration.