






Grey/White classification
This procedure yields a grey/white classification of each hemisphere for people wishing to test the influence of classification algorithms (SPM, MNI's tools, SHFJ's tools...) on the final VBM result.
The proposed method deals with each hemisphere individually, in order to prevent mixing between the two internal faces or between low cortex faces and cerebellum, during the spatial smoothing performed before statistics.
Another feature of the proposed approach: we have decided to discard cerebellum because partial volume effects are in our opinion too important to reach a correct binary classification (a fuzzy classification might overcome this problem).
mri_corrected: T1 MRI Bias Corrected ( input )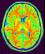
histo_analysis: Histo Analysis ( input )
split_mask: Split Brain Mask ( input )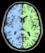
edges: T1 MRI Edges ( input )
commissure_coordinates: Commissure coordinates ( input )
Side: Choice ( input )Left, Right or Both Hemispheres
left_grey_white: Left Grey White Mask ( output )classification (grey=100, white=200)
right_grey_white: Right Grey White Mask ( output )classification (grey=100, white=200)
fix_random_seed: Boolean ( input )
Toolbox : Morphologist
User level : 2
Identifier :
GreyWhiteClassificationSupported file formats :
mri_corrected :GIS image, VIDA image, NIFTI-1 image, MINC image, gz compressed MINC image, DICOM image, TIFF image, XBM image, PBM image, PGM image, BMP image, XPM image, PPM image, gz compressed NIFTI-1 image, TIFF(.tif) image, ECAT i image, PNG image, JPEG image, MNG image, GIF image, SPM image, ECAT v imagehisto_analysis :Histo Analysissplit_mask :GIS image, VIDA image, NIFTI-1 image, MINC image, gz compressed MINC image, DICOM image, TIFF image, XBM image, PBM image, PGM image, BMP image, XPM image, PPM image, gz compressed NIFTI-1 image, TIFF(.tif) image, ECAT i image, PNG image, JPEG image, MNG image, GIF image, SPM image, ECAT v imageedges :GIS image, VIDA image, NIFTI-1 image, MINC image, gz compressed MINC image, DICOM image, TIFF image, XBM image, PBM image, PGM image, BMP image, XPM image, PPM image, gz compressed NIFTI-1 image, TIFF(.tif) image, ECAT i image, PNG image, JPEG image, MNG image, GIF image, SPM image, ECAT v imagecommissure_coordinates :Commissure coordinatesleft_grey_white :GIS image, VIDA image, NIFTI-1 image, MINC image, TIFF image, XBM image, PBM image, PGM image, BMP image, XPM image, PPM image, gz compressed NIFTI-1 image, ECAT i image, PNG image, JPEG image, MNG image, GIF image, SPM image, ECAT v imageright_grey_white :GIS image, VIDA image, NIFTI-1 image, MINC image, TIFF image, XBM image, PBM image, PGM image, BMP image, XPM image, PPM image, gz compressed NIFTI-1 image, ECAT i image, PNG image, JPEG image, MNG image, GIF image, SPM image, ECAT v image