Module: Molecular Diffusivity Experiment Simulation ()
WARNING: this feature is only available for Windows platform.
The Molecular Diffusivity Experiment Simulation module simulates an experiment leading to the computation of molecular diffusivity.
Note: see about system requirements and hardware platform availability.
Aknowledgments
This module was developed in collaboration with Dominique Bernard, Research Director at ICMCB-CNRS (Pessac, France).
Theoritical details
Some general elements about molecular diffusivity are exposed in . More precisely, the Fick's second equation is solved by imposing boundary conditions:
- one voxel wide plane of solid phase with null diffusion coefficient is added on the faces of the image that are not perpendicular to the main direction of the species transport. This allows isolating the sample from the outside.
- reservoirs with imposed initial concentrations are added at the input and output (the faces that are perpendicular to the flux main direction). They are designed so that the concentration in each reservoir is uniform.
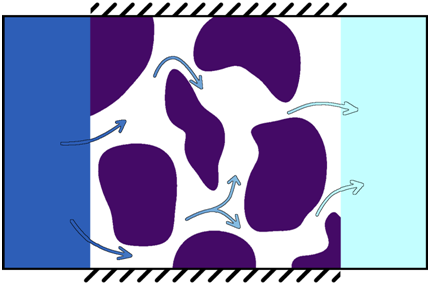
Physical constraints
Only one fluid phase and one solid phase are considered for this calculation: the module takes a label image as input, but each label of the image has to be part either of the solid phase or of the fluid phase. The diffusion coefficient of the solid phase is assumed to be zero. The rate of reaction of the solid is also assumed to be zero: there is no reaction occuring at the fluid-solid interface. The fluid phase is filled with a solvent and there is no flow. The diffusion of only one species is considered.
Computational aspects and results
As the computation can be rather long, it is left to the user to select whether one or several experiments must be computed. The results are presented in a spreadsheet with suffix .DExp.Spreadsheet.
There is one table for each direction (X, Y and Z). Each line of the spreadsheet represents a single computation of the properties. The columns contain:
- Geometry file: name of the file on which the computation was done,
- Region of interest: bounding box of the ROI on which the computation was performed,
- Apparent molecular diffusivity: value of the molecular diffusivity of the sample in [
],
- Input concentration: input concentration in [
] imposed at the entrance of the experimental setup,
- Output concentration: output concentration in [
] imposed at the exit of the experimental setup,
- Bulk solution diffusivity: molecular diffusivity of the free solution in [
].
Units and dimensions
The input and output concentration boundary conditions are supposed to be in [
]. The bulk solution diffusivity is in [
].
Problems and solutions
What does this error dialog mean?

Figure 1: Example of error dialog at the end of a computation. This dialog usually appears at the end of a computation and means that something went wrong during the computation. The solver did not reach the convergence target in the indicated number of iterations. There can be mainly two reasons to explain that problem.
First, the number of iterations is not large enough. It can be detected when the performed number of iterations equals the maximal number set as parameter. The maximal limit should be increased in the parameters of the modules. For large data volumes, the default value might be too small.
The other reason is more difficult to identify. When the discretization of the volume is too rough, the solver can start oscillating locally. One value is locally perturbed and each iteration modifies it consequently with respect to its absolute value. This value can be very small but the error is computed on relative variation between two time steps. For example, a value of
at iteration number 1 and
at iteration 2 varied by
between these two iterations. The value is very small, probably negligible for the final result, but the error remains large.
Several solutions can be tested to address these issues:
- Try to remove the non-percolating porosity from the input image. Non-percolating porosity is not involved in the physical property, but computation in it can be rather long to finally end to 0. To remove this porosity, the Axis Connectivity module can be used. The can help to apply this processing.
- Reduce the convergence criterion value, so that the solver iterates longer and reaches a lower error value. Also increase the maximum number of iteration, so that the solver is not bound by that number.
- Try to increase the refining coefficient. This parameter is hidden by default in the XLab modules parameters. To modify it, the Advanced Settings checkbox must be selected, then a slider appears below the list of materials. Setting refining coefficient to 2 means that all voxels is divided by 2 in the three directions of space. On one hand, it multiplies the number of unknowns to compute (probably memory consumption and computation time also) by a factor of 8. On the other hand, it increases the numeric precision and often helps the solver to converge when it could not with a refining coefficient of 1.
Data [required]
The input must be a Label Field.ROI [optional]
This is a Region Of Interest, meaning that the computation will only take into account the volume contained in the ROI.Initial Concentration Field [optional]
The connected field is used as initial solution of the problem. It means that the result of a previous computation can be reused to restart this computation from where it ended. It can be useful if the computation was interrupted before the targeted error value was reached. The connected field must have the same dimensions and voxel size as the data set. Otherwise, it will not be used for initializing the solution.
Options
This checkbox allows overwriting (checked) / creating new outputs (unchecked) when a computation ends.Pore Space
This port lists all the materials or labels contained in the connected Label Field. It allows selecting which labels or values must be considered as fluid for the computation. By default, the material named Exterior or the value 0 is selected.Iterations
Only appears when "show advanced settings" from port Options is checked. It is important to know that an explicit resolution is used to solve the Fick's equation system for the diffusivity computation. There are two numbers to set in this port. The first is the minimum number of iterations to compute. It is used to avoid ending the computation loop too early. It could happen in very specific cases, due to numerical oscillation in the first iterations. These oscillations can sometimes make the time derivative falsely lower than the convergence criterion. The oscillations should not remain beyond 500 iterations.Anyways, an option is activated by default to automatically estimate the minimum number of iterations necessary to make sure that the numerical oscillation of the first iterations is passed. In that case, the "min" entry is disabled and the number it contains is not taken into account. If the convergence criterion has not been set small enough, the computation will continue event if the criterion is reached, until the evaluated minimum number of iterations has been done.
The option can be deactivated by unchecking the "estimate min number" checkbox.
The estimation is done using Weissberg equation, which links the apparent diffusion coefficient to the porosity of the material. This estimate of the apparent diffusion coefficient gives a guess on the characteristic time of the experiment. This time can be used to obtain a number of iterations for which the initial oscillation is surely behind.
The second number refers to the maximum number of iterations to compute. It is used to be sure that the computation loop will end even if the convergence criterion cannot be attained because of numerical approximation problems. Each number can be set to a default value that should be enough for most cases. A computation should not need more than 100 000 iterations, which already is a huge value.
Transport Direction
This option allows the user to define the main direction of the species transport. Separate computations will be performed for each direction specified.Spreadsheet
This port selects where the next result must appear. If "append result" is selected, the next result is appended in the connected spreadsheet to the last created table corresponding to the species transport direction of this computation. If "new spreadsheet" is selected, the result is stored in a new spreadsheet. If no spreadsheet is connected, a new one is created.Outputs
If one of the check boxes is selected, the corresponding output will appear in the Project View at the end of the computation. If the first box is checked, a spreadsheet with suffix .DExp.ResConcentration.Spreadsheet is generated where the uniform concentrations of the input and output reservoirs are dumped at each iteration. A new table is opened for each new computation.The second check box stands for the resulting concentration field.
The third check box is checked by default. If the box is checked, a spreadsheet with suffix .DExp.Error.Spreadsheet is added to the Project View. It contains the estimation of the error (or convergence criterion) at each iteration, for each computation (depending on how many directions are selected).
Note: the concentration field is automatically created as output of the module if the computation was aborted or did not converge. The created field can be connected as an input of the module, so that the computed values can be used as initial solution of the problem (see above for more details).
Boundary Values
Input and output concentration boundary values imposed at the entrance and exit of the experimental setup. Those values are supposed to ne in []. They must be different, otherwise no diffusion occurs. If they are set to the same value, output concentration is automatically incremented by one.
Note: molecular diffusivity is independent from boundary conditions. Only the concentration field is rescaled according to boundary values.
Solution
Value of the molecular diffusivity of the free solution used for the experiment simulation, in [].
Note: molecular diffusivity is independent from boundary conditions. Only the concentration field is rescaled according to bulk solution diffusivity. However, lower values may increase the computation time, since diffusion will be slower.
hxportgroup Advanced Settings
Advanced Settings
This port sets whether the additional options for fine tuning the module are visible (ON) or hidden (OFF). The options that appear when checking are considered to be expert options, which should not require modification in most cases.
Refining Coefficient
Only appears when "show advanced settings" from port Options is checked. The maximum number of iterations to compute is used to be sure that the computation loop will end even if the convergence criterion cannot be attained because of numerical approximation problems.Convergence Criterion
Only appears when "show advanced settings" from port Options is checked. It is assumed that convergence is attained when the time derivatives of the unknowns tend to 0. Numerically, 0 can never be reached; that is why a precision value must be set to indicate that the convergence is sufficient. This is a floating point value which must be greater than 0 and for which a default value of 10is suggested.
Warning: the default value is not appropriate for any computation. Depending on the geometry of the sample, it might be mandatory to decrease the precision value drastically.
For convenience, the convergence criterion will often be simply called "error" in the GUI, spreadsheets and Console messages.
Expose
This checkbox only appears when "advanced settings" is ON. It stands for the refined image. The output that is shown in the Project View is the refined subvolume used for the computation. It means that the output field fits in the ROI if it is defined or has the same dimensions as the input data. It also means that the output field is oversampled by the factor indicated in the refining coefficient option.