Parallel Computing Toolbox Release Notes
Discontinued support for parallel computing products on 32-bit Windows operating systems
This release of MATLAB® products no longer supports 32-bit Parallel Computing Toolbox™ and MATLAB Distributed Computing Server™ on Windows® operating systems.
Compatibility Considerations
You can no longer install the parallel computing products on 32-bit Windows operating systems. If you must use Windows operating systems for the parallel computing products, upgrade to 64-bit MATLAB products on a 64-bit operating system.
More than 90 GPU-enabled functions in Statistics and Machine Learning Toolbox, including probability distribution, descriptive statistics, and hypothesis testing
Statistics and Machine Learning Toolbox™ offers enhanced functionality for some of its functions to perform computations on a GPU. For specific information, see the Statistics and Machine Learning Toolbox release notes. Parallel Computing Toolbox is required to access this functionality.
Additional GPU-enabled MATLAB functions, including support for sparse matrices
Additional GPU-enabled MATLAB functions
The following functions are new in their support for gpuArrays:
assert bandwidth deg2rad expm gmres intersect | isbanded rad2deg randperm rectint repelem setdiff | setxor sortrows union unique |
In addition to the functions above, the following functions are enhanced in their gpuArray support:
arrayfunnow supports code that includes the functionsisfloat,isinteger,islogical,isnumeric,isreal, andissparse.pagefunsupport for@mldivideand@mrdivideis no longer limited to 32-by-32 pages.cov,max,mean,median,min,std,sum, andvarnow support the'omitnan'option.accumarraynow supports the logical sparse argument, for generating a sparse gpuArray output.
For a list of MATLAB functions that support gpuArray, see Run Built-In Functions on a GPU .
Sparse arrays with GPU-enabled functions
The following functions are new in their support for sparse gpuArrays:
abs angle ceil deg2rad expm1 fix floor gmres | isequal isequaln log1p minus nextpow2 plus rad2deg realsqrt | round sign spfun sqrt sum uminus uplus |
You can create a sparse gpuArray either by calling sparse with a gpuArray input, or by
calling gpuArray with a sparse
input.
For more information on this topic, see Sparse Arrays on a GPU.
mexcuda function for easier compilation of MEX-files containing CUDA code
A new mexcuda function allows you to compile
MEX-functions for GPU computation.
For more information, see the mexcuda function
reference page. See also Run CUDA or PTX Code on GPU.
Upgraded CUDA Toolkit version
The parallel computing products are now using CUDA® Toolkit version 7.0. To compile CUDA code for CUDAKernel or CUDA MEX-files, you must use toolkit version 7.0.
Scheduler integration scripts for SLURM
This release offers a new set of scripts containing submit and decode functions to support Simple Linux® Utility for Resource Management (SLURM), using the generic scheduler interface. The pertinent code files are in the folder:
matlabroot/toolbox/distcomp/examples/integration/slurm
where matlabroot is your installation
location.
The slurm folder contains a README file
of instructions, and folders for shared, nonshared,
and remoteSubmission network configurations.
For more information, view the files in the appropriate folders. See also Program Independent Jobs for a Generic Scheduler and Program Communicating Jobs for a Generic Scheduler.
parallel.pool.Constant function to create constant data on parallel pool workers, accessible within parallel
language constructs such as parfor and parfeval
A new parallel.pool.Constant function allows
you to define a constant whose value can be accessed by multiple parfor-loops
or other parallel language constructs (e.g., spmd or parfeval)
without the need to transfer the data multiple times.
For more information and examples, see parallel.pool.Constant.
Improved performance of mapreduce on Hadoop 2 clusters
The performance of mapreduce running on
a Hadoop® 2.x cluster with MATLAB Distributed Computing Server is
improved in this release for large input data.
Enhanced and additional MATLAB functions for distributed arrays
The following functions are new in supporting distributed arrays, with all forms of codistributor (1-D and 2DBC):
bicg deg2rad polyarea polyint | rad2deg rectint repelem |
In addition to the new functions above, the following functions are enhanced in their distributed array support:
cov,max,mean,median,min,std,sum, andvarnow support the'omitnan'option.
For a list of MATLAB functions that support distributed arrays, see MATLAB Functions on Distributed and Codistributed Arrays.
Warnings property for tasks
There is a new Warnings property for parallel.Task objects. This property
contains warning information that was issued during execution of the
task. The data type is a structure array with the fields message, identifier,
and stack. This information is part of the job display in the Command Window
and Job Monitor.
More consistent transparency enforcement
For more consistent behavior and results, transparency violations
in parfor-loops and spmd blocks
are now being enforced more strictly.
Compatibility Considerations
It is possible that parfor or spmd code
that did not error in previous releases will now generate a transparency
violation error. For more information, see Transparency
.
Anonymous functions that were saved in a previous release and that reference nested functions will not execute in this release. They will load, but when executed will issue an "Undefined function handle" error.
Upgrade parallel computing products together
This version of Parallel Computing Toolbox software is accompanied by a corresponding new version of MATLAB Distributed Computing Server software.
Compatibility Considerations
As with every new release, if you are using both parallel computing products, you must upgrade Parallel Computing Toolbox and MATLAB Distributed Computing Server together. These products must be the same version to interact properly with each other.
Jobs created in one version of Parallel Computing Toolbox software
will not run in a different version of MATLAB Distributed Computing Server software,
and might not be readable in different versions of the toolbox software.
The job data stored in the folder identified by JobStorageLocation (formerly DataLocation)
might not be compatible between different versions of MATLAB Distributed Computing Server.
Therefore, JobStorageLocation should not be shared
by parallel computing products running different versions, and each
version on your cluster should have its own JobStorageLocation.
Support for mapreduce function on any cluster that supports parallel pools
You can now run parallel mapreduce on
any cluster that supports a parallel pool. For more information, see Run
mapreduce on a Parallel Pool.
Sparse arrays with GPU-enabled functions
This release supports sparse arrays on a GPU. You can create
a sparse gpuArray either by calling sparse with
a gpuArray input, or by calling gpuArray with
a sparse input. The following functions support sparse gpuArrays.
classUnderlying conj ctranspose end find full gpuArray.speye imag isaUnderlying isempty | isfloat isinteger islogical isnumeric isreal issparse length mtimes ndims nonzeros | nnz numel nzmax real size sparse spones transpose |
Note the following for some of these functions:
gpuArray.speyeis a static constructor method.sparsesupports only single-argument syntax.mtimesdoes not support the case of full-matrix times a sparse-matrix.
For more information on this topic, see Sparse Arrays on a GPU.
A new C function, mxGPUIsSparse,
is available for the MEX interface, to query whether a gpuArray is
sparse or not. However, even though the MEX interface can query properties
of a sparse gpuArray, its functions cannot access sparse gpuArray
elements.
Additional GPU-enabled MATLAB functions
The following functions are new in their support for gpuArrays:
cdf2rdf gpuArray.freqspace histcounts idivide inpolygon isdiag ishermitian issymmetric | istril istriu legendre nonzeros nthroot pinv planerot poly | polyarea polyder polyfit polyint polyval polyvalm |
Note the following for some of these functions:
gpuArray.freqspaceis a static constructor method.
For a list of MATLAB functions that support gpuArray, see Run Built-In Functions on a GPU.
pagefun support for mrdivide and inv functions on GPUs
For gpuArray inputs, pagefun is
enhanced to support:
@inv@mrdivide(for square matrix divisors of sizes up to 32-by-32)
Enhancements to GPU-enabled linear algebra functions
Many of the linear algebra functions that support gpuArrays
are enhanced for improved performance. Among those functions that
can exhibit improved performance are svd, null, eig (for
nonsymmetric input), and mtimes (for inner products).
Parallel data reads from a datastore with MATLAB partition function
The partition function can perform a parallel
read and partition of a Datastore. For
more information, see partition and numpartitions.
See also Partition
a Datastore in Parallel.
Using DNS for cluster discovery
In addition to multicast, the discover cluster functionality of Parallel Computing Toolbox can now use DNS to locate MATLAB job scheduler (MJS) clusters. For information about cluster discovery, see Discover Clusters. For information about configuring and verifying the required DNS SRV record on your network, see DNS SRV Record.
MS-MPI support for local and MJS clusters
On 64-bit Windows platforms, Microsoft® MPI (MS-MPI) is now the default MPI implementation for local clusters on the client machine.
For MATLAB job scheduler (MJS) clusters on Windows platforms,
you can use MS-MPI by specifying the -useMSMPI flag
with the startjobmanager command.
Ports and sockets in mdce_def file
The following parameters are new to the mdce_def file
for controlling the behavior of MATLAB job scheduler (MJS) clusters.
ALL_SERVER_SOCKETS_IN_CLUSTER— This parameter controls whether all client connections are outbound, or if inbound connections are also allowed.JOBMANAGER_PEERSESSION_MIN_PORT,JOBMANAGER_PEERSESSION_MAX_PORT— These parameters set the range of ports to use whenALL_SERVER_SOCKETS_IN_CLUSTER=true.WORKER_PARALLELPOOL_MIN_PORT,WORKER_PARALLELPOOL_MAX_PORT— These parameters set the range of ports to use on worker machines for parallel pools.
For more information and default settings for these parameters,
see the appropriate mdce_def file for your platform:
matlabroot\toolbox\distcomp\bin\mdce_def.batmatlabroot/toolbox/distcomp/bin/mdce_def.sh
Compatibility Considerations
By default in this release, ALL_SERVER_SOCKETS_IN_CLUSTER is true,
which makes all connections outbound from the client. For pre-R2015a
behavior, set its value to false, which also initiates
a set of inbound connections to the client from the MJS and workers.
Improved profiler accuracy for GPU code
The MATLAB profiler now reports more accurate timings for code running on a GPU. For related information, see Measure Performance on the GPU.
Upgraded CUDA Toolkit version
The parallel computing products are now using CUDA Toolkit version 6.5. To compile CUDA code for CUDAKernel or CUDA MEX files, you must use toolkit version 6.5.
Discontinued support for GPU devices on 32-bit Windows computers
This release no longer supports GPU devices on 32-bit Windows machines.
Compatibility Considerations
GPU devices on 32-bit Windows machines are not supported in this release. Instead, use GPU devices on 64-bit machines.
Discontinued support for parallel computing products on 32-bit Windows computers
In a future release, support will be removed for Parallel Computing Toolbox and MATLAB Distributed Computing Server on 32-bit Windows machines.
Compatibility Considerations
Parallel Computing Toolbox and MATLAB Distributed Computing Server are still supported on 32-bit Windows machines in this release, but parallel language commands can generate a warning. In a future release, support will be completely removed for these computers, at which time it will not be possible to install the parallel computing products on them.
matlabpool function removed
The matlabpool function has been removed.
Compatibility Considerations
Calling matlabpool now generates an error.
You should instead use parpool to
create a parallel pool.
Parallelization of mapreduce on local workers
If you have Parallel Computing Toolbox installed, and your
default cluster profile specifies a local cluster, then execution
of mapreduce opens
a parallel pool and distributes tasks to the pool workers.
Note
If your default cluster profile specifies some other cluster,
the |
For more information, see Run mapreduce on a Local Cluster.
Additional GPU-enabled MATLAB functions, including accumarray, histc, cummax, and cummin
The following functions are new in their support for gpuArrays:
accumarray acosd acotd acscd asecd asind atan2d atand betainc betaincinv cart2pol cart2sph compan corrcoef cosd cotd cscd | cummax cummin del2 factorial gammainc gammaincinv gradient hankel histc hsv2rgb isaUnderlying median mode nextpow2 null orth pol2cart | psi rgb2hsv roots secd sind sph2cart subspace superiorfloat swapbytes tand toeplitz trapz typecast unwrap vander |
Note the following for some of these functions:
The first input argument to
gammainccannot contain any negative elements.
For a list of MATLAB functions that support gpuArray, see Run Built-In Functions on a GPU.
pagefun support for mldivide on GPUs
For gpuArray inputs, pagefun is
enhanced to support @mldivide for square matrix
divisors of sizes up to 32-by-32.
Additional MATLAB functions for distributed arrays, including fft2, fftn, ifft2, ifftn, cummax, cummin, and diff
The following functions now support distributed arrays with all forms of codistributor (1-D and 2DBC), or are enhanced in their support for this release:
besselh besseli besselj besselk bessely beta betainc betaincinv betaln cart2pol cart2sph compan corrcoef cov cummax cummin diff | erf erfc erfcinv erfcx erfinv fft2 fftn gamma gammainc gammaincinv gammaln hankel hsv2rgb ifft ifft2 ifftn isfloat | isinteger islogical isnumeric median mode pol2cart psi rgb2hsv sph2cart std toeplitz trapz unwrap vander var |
Note the following for some of these functions:
isfloat,isinteger,islogical, andisnumericnow return results based onclassUnderlyingof the distributed array.
For a list of MATLAB functions that support distributed arrays, see MATLAB Functions on Distributed and Codistributed Arrays.
Data Analysis on Hadoop clusters using mapreduce
Parallel Computing Toolbox and MATLAB Distributed Computing Server support
the use of Hadoop clusters for the execution environment of mapreduce applications.
For more information, see:
Discover Clusters Supports Microsoft Windows HPC Server for Multiple Releases
The Discover Clusters dialog can now list Microsoft Windows HPC Server clusters for different releases of parallel computing products. For more information about cluster discovery, see Discover Clusters.
For this functionality, the HPC Server client utilities must be installed on the client machine from which you are discovering. See Configure Client Computer for HPC Server.
Upgraded CUDA Toolkit Version
The parallel computing products are now using CUDA Toolkit version 6.0. To compile CUDA code for CUDAKernel or CUDA MEX files, you must use toolkit version 6.0 or earlier.
Discontinued Support for GPU Devices of Compute Capability 1.3
This release no longer supports GPU devices of compute capability 1.3.
Compatibility Considerations
This release supports only GPU devices of compute capability 2.0 or greater.
Discontinued Support for GPU Devices on 32-Bit Windows Computers
In a future release, support for GPU devices on 32-bit Windows machines will be removed.
Compatibility Considerations
GPU devices on 32-bit Windows machines are still supported in this release, but in a future release support will be completely removed for these devices.
Number of local workers no longer limited to 12
You can now run a local cluster of more than 12 workers on your client machine. Unless you adjust the cluster profile, the default maximum size for a local cluster is the same as the number of computational cores on the machine.
Additional GPU-enabled MATLAB functions: interp3, interpn, besselj, bessely
The following functions are new in their support for gpuArrays:
besselj bessely interp3 interpn |
For a list of MATLAB functions that support gpuArray, see Run Built-In Functions on a GPU.
Additional GPU enabled Image Processing Toolbox functions: bwdist, imreconstruct, iradon, radon
Image Processing Toolbox™ offers enhanced functionality for some of its functions to perform computations on a GPU. For specific information, see the Image Processing Toolbox release notes. Parallel Computing Toolbox is required to access this functionality.
Enhancements to GPU-enabled MATLAB functions: filter (IIR filters); pagefun (additional functions supported); interp1, interp2, conv2, reshape (performance
improvements)
The following functions are enhanced in their support for gpuArray data:
conv2 filter interp1 interp2 pagefun | rand randi randn reshape |
Note the following enhancements for some of these functions:
filternow supports IIR filtering.pagefunis enhanced to support most element-wise gpuArray functions. Also, these functions are supported:@ctranspose,@fliplr,@flipud,@mtimes,@rot90,@transpose.rand(___,'like',P)P. This enhancement also applies torandi,randn.
For a list of MATLAB functions that support gpuArray, see Run Built-In Functions on a GPU.
Duplication of an existing job, containing some or all of its tasks
You can now duplicate job objects, allowing you to resubmit jobs that had finished or failed.
The syntax to duplicate a job is
newjob = recreate(oldjob)
where oldjob is an existing job object. The newjob object
has all the same tasks and settable properties as oldjob,
but receives a new ID. The old job can be in any
state; the new job state is pending.
You can also specify which tasks from an existing independent job to include in the new job, based on the task IDs. For example:
newjob = recreate(oldjob,'TaskID',[33:48]);For more information, see the recreate reference
page.
More MATLAB functions enhanced for distributed arrays
The following functions now support distributed arrays with all forms of codistributor (1-D and 2DBC), or are enhanced in their support for this release:
eye ifft rand randi randn |
Note the following enhancements for some of these functions:
For a list of MATLAB functions that support distributed arrays, see MATLAB Functions on Distributed and Codistributed Arrays.
GPU MEX Support for Updated MEX
The GPU MEX support for CUDA code now incorporates the latest MATLAB MEX functionality. See Streamlined MEX compiler setup and improved troubleshooting.
Compatibility Considerations
The latest MEX does not support the mexopts files
shipped in previous releases. Updated .xml files
are provided instead. To use the updated MEX functionality and to
avoid a warning, replace the mexopts file you used
in past releases with the appropriate new .xml file
as described in Set
Up for MEX-File Compilation.
Old Programming Interface Removed
The programming interface characterized by distributed jobs
and parallel jobs has been removed. This old interface used functions
such as findResource, createParallelJob, getAllOutputArguments, dfeval,
etc.
Compatibility Considerations
The functions of the old programming interface now generate errors. You must migrate your code to the interface described in the R2012a release topic New Programming Interface.
matlabpool Function Being Removed
The matlabpool function is being removed.
Compatibility Considerations
Calling matlabpool continues to work in this
release, but now generates a warning. You should instead use parpool to
create a parallel pool.
Removed Support for parallel.cluster.Mpiexec
Support for clusters of type parallel.cluster.Mpiexec has been removed.
Compatibility Considerations
Any attempt to use parallel.cluster.Mpiexec clusters now generates an error. As an alternative, consider using the generic scheduler interface, parallel.cluster.Generic.
parpool: New command-line interface (replaces matlabpool), desktop indicator, and preferences for easier interaction with a parallel
pool of MATLAB workers
Parallel Pool
Replaces MATLAB Pool
Parallel pool syntax replaces MATLAB pool syntax for executing
parallel language constructs such as parfor, spmd, Composite,
and distributed. The pool is represented in MATLAB
by a parallel.Pool object.
The general workflow for these parallel constructs remains the
same: When the pool is available, parfor, spmd,
etc., run the same as before. For example:
| MATLAB Pool | Parallel Pool |
|---|---|
matlabpool open 4
parfor ii=1:n
X(n) = myFun(n);
end
matlabpool close | p = parpool(4)
parfor ii=1:n
X(n) = myFun(n);
end
delete(p) |
Functions for Pool Control
The following functions provide command-line interface for controlling a parallel pool. See the reference pages for more information about each.
| Function | Description |
|---|---|
parpool | Start a parallel pool |
gcp | Get current parallel pool or start pool |
delete | Shut down and delete the parallel pool |
addAttachedFiles | Attach files to the parallel pool |
listAutoAttachedFiles | List files automatically attached to the parallel pool |
updateAttachedFiles | Send updates to files already attached to the parallel pool |
Asynchronous Function Evaluation on Parallel Pool
You can evaluate functions asynchronously on one or all workers
of a parallel pool. Use parfeval to evaluate
a function on only one worker, or use parfevalOnAll to
evaluate a function on all workers in the pool.
parfeval or parfevalOnAll returns
an object called a future, from which you can
get the outputs of the asynchronous function evaluation. You can create
an array of futures by calling parfeval in a for-loop,
setting unique parameters for each call in the array.
The following table lists the functions for submitting asynchronous evaluations to a parallel pool, retrieving results, and controlling future objects. See the reference page of each for more details.
| Function | Description |
|---|---|
parfeval | Evaluate function asynchronously on worker in parallel pool |
parfevalOnAll | Evaluate function asynchronously on all workers in parallel pool |
fetchOutputs | Retrieve all output arguments from future |
fetchNext | Retrieve next available unread future outputs |
cancel | Cancel queued or running future |
wait | Wait for future to complete |
For more information on parallel future objects, including their methods and properties, see the parallel.Future reference page.
Compatibility Considerations
This release continues to support MATLAB pool language usage, but this support might discontinue in future releases. You should update your code as soon as possible to use parallel pool syntax instead.
New Desktop Pool Indicator
A new icon at the lower-left corner of the desktop indicates the current pool status.
![]()
Icon color and tool tips let you know if the pool is busy or ready, how large it is, and when it might time out. You can click the icon to start a pool, stop a pool, or access your parallel preferences.
New Parallel Preferences
Your MATLAB preferences now include a group of settings for parallel preferences. These settings control general behavior of clusters and parallel pools for your MATLAB session.
You can access your parallel preferences in these ways:
In the Environment section of the Home tab, click Parallel > Parallel Preferences
Click the desktop pool indicator icon, and select Parallel preferences.
Type
preferencesat the command line, and click the Parallel Computing Toolbox node.

Settings in your parallel preferences control the default cluster choice, and the preferred size, automatic opening, and timeout conditions for parallel pools. For more information about these settings, see Parallel Preferences.
Automatic start of a parallel pool when executing code that uses parfor or spmd
You can set your parallel preferences so that a parallel pool
automatically starts whenever you execute a language construct that
runs on a pool, such as parfor, spmd, Composite, distributed, parfeval,
and parfevalOnAll.
Compatibility Considerations
The default preference setting is to automatically start a pool
when a parallel language construct requires it. If you want to make
sure a pool does not start automatically, you must change your parallel
preference setting. You can also work around this by making sure to
explicitly start a parallel pool with parpool before
encountering any code that needs a pool.
By default, a parallel pool will shut down if idle for 30 minutes. To prevent this, change the setting in your parallel preferences; or the pool indicator tool tip warns of an impending timeout and provides a link to extend it.
Option to start a parallel pool without using MPI
You now have the option to start a parallel pool on a local
or MJS cluster so that the pool does not support running SPMD constructs.
This allows the parallel pool to keep running even if one or more
workers aborts during parfor execution. You explicitly
disable SPMD support when starting the parallel pool by setting its 'SpmdEnabled' property false in
the call to the parpool function. For example:
p = parpool('SpmdEnabled',false);Compatibility Considerations
Running any code (including MathWorks toolbox code) that uses SPMD constructs, on a parallel pool that was created without SPMD support, will generate errors.
More GPU-enabled MATLAB functions (e.g., interp2, pagefun) and Image Processing Toolbox functions (e.g., bwmorph, edge, imresize,
and medfilt2)
Image Processing Toolbox offers enhanced functionality for some of its functions to perform computations on a GPU. For specific information, see the Image Processing Toolbox release notes. Parallel Computing Toolbox is required to access this functionality.
The following functions are new or enhanced in Parallel Computing Toolbox to support gpuArrays:
flip interp2 pagefun
Note the following for some of these functions:
pagefunallows you to iterate over the pages of a gpuArray, applying@mtimesto each page.For more information, see the
pagefunreference page, or typehelp pagefun.
For complete lists of functions that support gpuArray, see Run Built-In Functions on a GPU.
More MATLAB functions enabled for distributed arrays: permute, ipermute, and sortrows
The following functions now support distributed arrays with all forms of codistributor (1-D and 2DBC), or are enhanced in their support for this release:
ipermute permute sortrows cast | zeros ones nan inf true false |
Note the following enhancements for some of these functions:
ipermute,permute, andsortrowssupport distributed arrays for the first time in this release.castsupports the'like'option for distributed arrays, applying the underlying class of one array to another.Z = zeros(___,'like',P)returns a distributed array of zeros of the same complexity as distributed arrayP, and same underlying class asPif class is not specified in the function call. The same behavior applies to the other similar constructors in the right-hand column of this table.
For more information on any of these functions, type help
distributed.. For
example:functionname
help distributed.ipermute
For complete lists of MATLAB functions that support distributed arrays, see MATLAB Functions on Distributed and Codistributed Arrays.
Enhancements to MATLAB functions enabled for GPUs, including ones, zeros
The following functions are enhanced in their support for gpuArray data:
zeros ones nan inf | eye true false cast |
Note the following enhancements for these functions:
Z = zeros(___,'like',P)returns a gpuArray of zeros of the same complexity as gpuArrayP, and same underlying class asPif class is not specified. The same behavior applies to the other constructor functions listed in this table.castalso supports the'like'option for gpuArray input, applying the underlying class of one array to another.
For more information on any of these functions, type help
gpuArray.. For example:functionname
help gpuArray.cast
For complete lists of MATLAB functions that support gpuArray, see Run Built-In Functions on a GPU.
New GPU Random Number Generator NormalTransform Option: Box-Muller
When generating random numbers on a GPU, there is a new option
for 'NormalTransform' called 'BoxMuller'.
The Box-Muller transform allows faster generation of normally distributed
random numbers on the GPU.
This new option is the default 'NormalTransform' setting
when using the Philox4x32-10 or Threefry4x64-20 generator.
The following commands, therefore, use 'BoxMuller' for 'NormalTransform':
parallel.gpu.rng(0,'Philox4x32-10') parallel.gpu.rng(0,'Threefry4x64-20')
Note
The |
Compatibility Considerations
In previous releases, the default 'NormalTransform' setting
when using the Philox4x32-10 or Threefry4x64-20 generator
on a GPU was 'Inversion'. If you used either of
these generators with the default 'NormalTranform' and
you want to continue with the same behavior, you must explicitly set
the 'NormalTransform' with either of these commands:
stream = parallel.gpu.RandStream('Philox4x32-10','NormalTransform','Inversion')
parallel.gpu.RandStream.setGlobalStream(stream)
stream = parallel.gpu.RandStream('Threefry4x64-20','NormalTransform','Inversion')
parallel.gpu.RandStream.setGlobalStream(stream)Upgraded MPICH2 Version
The parallel computing products are now shipping MPICH2 version 1.4.1p1 on all platforms.
Compatibility Considerations
If you use your own MPI builds, you might need to create new builds compatible with this latest version, as described in Use Different MPI Builds on UNIX Systems.
Discontinued Support for GPU Devices of Compute Capability 1.3
In a future release, support for GPU devices of compute capability 1.3 will be removed. At that time, a minimum compute capability of 2.0 will be required.
Compatibility Considerations
In R2013b, any use of gpuDevice to select
a GPU with compute capability 1.3, generates a warning. The device
is still supported in this release, but in a future release support
will be completely removed for these 1.3 devices.
GPU-enabled functions in Image Processing Toolbox and Phased Array System Toolbox
More toolboxes offer enhanced functionality for some of their functions to perform computations on a GPU. For specific information about these other toolboxes, see their respective release notes. Parallel Computing Toolbox is required to access this functionality.
More MATLAB functions enabled for use with GPUs, including interp1 and ismember
The following functions are enhanced to support gpuArray data:
interp1 isfloat isinteger | ismember isnumeric |
Note the following for some of these functions:
interp1supports only the linear and nearest interpolation methods.isfloat,isinteger, andisnumericnow return results based onclassUnderlyingof the gpuArray.ismemberdoes not support the'rows'or'legacy'option for gpuArray input.
For complete lists of functions that support gpuArray, see Built-In Functions That Support gpuArray.
Enhancements to MATLAB functions enabled for GPUs, including arrayfun, svd, and mldivide (\)
The following functions are enhanced in their support for gpuArray data:
arrayfun bsxfun mldivide | mrdivide svd |
Note the following enhancements for some of these functions:
arrayfunandbsxfunsupport indexing and accessing variables of outer functions from within nested functions.arrayfunsupports singleton expansion of all arguments for all operations. For more information, see thearrayfunreference page.mldivideandmrdividesupport all rectangular arrays.svdcan perform economy factorizations.
For complete lists of MATLAB functions that support gpuArray, see Built-In Functions That Support gpuArray.
Ability to launch CUDA code and manipulate data contained in GPU arrays from MEX-functions
You can now compile MEX-files that contain CUDA code, to create functions that support gpuArray input and output. This functionality is supported only on 64-bit platforms (win64, glnxa64, maci64). For more information and examples, see Execute MEX-Functions Containing CUDA Code.
For a list of C functions supporting this capability, see the group of C Functions in GPU Computing.
Automatic detection and transfer of files required for execution in both batch and interactive workflows
Parallel Computing Toolbox can now automatically attach files
to a job so that workers have the necessary code files for evaluating
tasks. When you set a job object's AutoAttachFiles to
true, an analysis determines what files on the client machine are
necessary for the evaluation of your job, and those files are automatically
attached to the job and sent to the worker machines.
You can set the AutoAttachFiles property
in the Cluster Profile Manager, or at the command-line. To get a listing
of the files that are automatically attached, use the listAutoAttachedFiles method
on a job or task object.
For more information, see Pass Data to and from Worker Sessions.
If the AutoAttachFiles property in the
cluster profile for the MATLAB pool is set to true, MATLAB performs
an analysis on spmd blocks
and parfor-loops
to determine what code files are necessary for their execution, then
automatically attaches those files to the MATLAB pool job so
that the code is available to the workers.
When you use the MATLAB editor to update files on the client
that are attached to a matlabpool,
those updates are automatically propagated to the workers in the
pool.
More MATLAB functions enabled for distributed arrays
The following functions now support distributed arrays with all forms of codistributor (1-D and 2DBC), or are enhanced in their support for this release:
cumprod cumsum eig | qr prod |
Note the following enhancements for some of these functions:
eigsupports generalized eigenvalues for symmetric matrices.qrsupports column pivoting.cumprod,cumsum, andprodnow support all integer data types; andprodaccepts the optional'native'or'double'argument.
More MATLAB functions enabled for GPUs, including convn, cov, and normest
gpuArray Support
The following functions are enhanced to support gpuArray data, or are expanded in their support:
bitget bitset cond convn | cov issparse mpower nnz | normest pow2 var |
The following functions are not methods of the gpuArray class, but they now work with gpuArray data:
blkdiag cross iscolumn | ismatrix isrow | isvector std |
For complete lists of functions that support gpuArray, see Built-In Functions That Support gpuArray.
MATLAB Code on the GPU
bsxfun now supports the same subset of
the language on a GPU that arrayfun does.
GPU support is extended to include the following MATLAB code
in functions called by arrayfun and bsxfun to
run on the GPU:
bitget bitset pow2 |
GPU-enabled functions in Neural Network Toolbox, Phased Array System Toolbox, and Signal Processing Toolbox
A number of other toolboxes now support enhanced functionality for some of their functions to perform computations on a GPU. For specific information about these other toolboxes, see their respective release notes. Parallel Computing Toolbox is required to access this functionality.
Performance improvements to GPU-enabled MATLAB functions and random number generation
The performance of some MATLAB functions and random number generation on GPU devices is improved in this release.
You now have a choice of three random generators on the GPU: the combined multiplicative recursive MRG32K3A, the Philox4x32-10, and the Threefry4x64-20. For information on these generators and how to select them, see Control the Random Stream for gpuArray. For information about generating random numbers on a GPU, and a comparison between GPU and CPU generation, see Control Random Number Streams.
Automatic detection and selection of specific GPUs on a cluster node when multiple GPUs are available on the node
When multiple workers run on a single compute node with multiple
GPU devices, the devices are automatically divided up among the workers.
If there are more workers than GPU devices on the node, multiple workers
share the same GPU device. If you put a GPU device in 'exclusive' mode,
only one worker uses that device. As in previous releases, you can
change the device used by any particular worker with the gpuDevice function.
More MATLAB functions enabled for distributed arrays, including sparse constructor, bsxfun,
and repmat
The following functions now support distributed arrays with all forms of codistributor (1-D and 2DBC), or are enhanced in their support for this release:
atan2d bsxfun cell2struct | mrdivide repmat sparse | struct2cell typecast |
Note the following enhancements for some of these functions:
cell2struct,struct2cell, andtypecastnow support 2DBC in addition to 1-D distribution.mrdivideis now fully supported, and is no longer limited to accepting only scalars for its second argument.sparseis now fully supported for all distribution types.
This release also offers improved performance of fft functions for long vectors as distributed arrays.
Detection of MATLAB Distributed Computing Server clusters that are available for connection from user desktops through Profile Manager
You can let MATLAB discover clusters for you. Use either of the following techniques to discover those clusters which are available for you to use:
On the Home tab in the Environment section, click Parallel > Discover Clusters.
In the Cluster Profile Manager, click Discover Clusters.
For more information, see Discover Clusters.
gpuArray Class Name
The object type formerly known as a GPUArray has been renamed
to gpuArray. The corresponding class name has been changed from parallel.gpu.GPUArray
to the shorter gpuArray. The name of the function gpuArray remains
unchanged.
Compatibility Considerations
You cannot load gpuArray objects from files that were saved in previous versions.
Code that uses the old class name must be updated to use the shorter new name. For example, the functions for directly generating gpuArrays on the GPU:
| Previous version form | New version form |
|---|---|
parallel.gpu.GPUArray.rand parallel.gpu.GPUArray.ones | gpuArray.rand gpuArray.ones |
Diary Output Now Available During Running Task
Diary output from tasks (including those of batch jobs) can
now be obtained while the task is still running. The diary is text
output that would normally be sent to the Command Window. Now this
text is appended to the task's Diary property
as the text is generated, rather than waiting until the task is complete.
You can read this property at any time. Diary information is accumulated
only if the job's CaptureDiary property
value is true. (Note:
This feature is not yet available for SOA jobs on HPC Server clusters.)
New Programming Interface
This release provides a new programming interface for accessing clusters, jobs, and tasks.
General Concepts and Phrases
This table maps some of the concepts and phrases from the old interface to the new.
| Previous Interface | New Interface in R2012a |
|---|---|
| MathWorks job manager | MATLAB job scheduler (MJS) |
| Third-party or local scheduler | Common job scheduler (CJS) |
| Configuration | Profile |
| Scheduler | Cluster |
The following code examples compare programming the old and new interfaces, showing some of the most common commands and properties. Note that most differences involve creating a cluster object instead of a scheduler object, and some of the property and method names on the job. After the example are tables listing some details of these new objects.
| Previous Interface | New Interface |
|---|---|
sched = findResource('scheduler',...
'Configuration','local');
j = createJob(sched,...
'PathDependencies',{'/share/app/'},...
'FileDependencies',{'funa.m','funb.m'};
createTask(j,@myfun,1,{3,4});
submit(j);
waitForState(j);
results = j.getAllOutputArguments; | clust = parcluster('local');
j = createJob(clust,...
'AdditionalPaths',{'/share/app/'}),...
'AttachedFiles,{'funa.m','funb.m'};
createTask(j,@myfun,1,{3,4});
submit(j);
wait(j);
results = j.fetchOutputs;
|
Objects
These tables compare objects in the previous and new interfaces.
| Previous Scheduler Objects | New Cluster Objects |
|---|---|
ccsscheduler | parallel.cluster.HPCServer |
genericscheduler | parallel.cluster.Generic |
jobmanager | parallel.cluster.MJS |
localscheduler | parallel.cluster.Local |
lsfscheduler | parallel.cluster.LSF |
mpiexec | parallel.cluster.Mpiexec |
pbsproscheduler | parallel.cluster.PBSPro |
torquescheduler | parallel.cluster.Torque |
For information on each of the cluster objects, see the parallel.Cluster reference
page.
| Previous Job Objects | New Job Objects |
|---|---|
job (distributed job) | parallel.job.MJSIndependentJob |
matlabpooljob | parallel.job.MJSCommunicatingJobwhere ( 'Type' = 'Pool') |
paralleljob | parallel.job.MJSCommunicatingJobwhere ( 'Type' = 'SPMD') |
simplejob | parallel.job.CJSIndependentJob |
simplematlabpooljob | parallel.job.CJSCommunicatingJob( 'Type'
= 'Pool') |
simpleparalleljob | parallel.job.CJSCommunicatingJob( 'Type'
= 'SPMD') |
For information on each of the job objects, see the parallel.Job reference
page.
| Previous Task Objects | New Task Objects |
|---|---|
task | parallel.task.MJSTask |
simpletask | parallel.task.CJSTask |
For information on each of the task objects, see the parallel.Task reference
page.
| Previous Worker Object | New Worker Objects |
|---|---|
worker | parallel.cluster.MJSWorker,parallel.cluster.CJSWorker |
For information on each of the worker objects, see the parallel.Worker reference
page.
Functions and Methods
This table compares some functions and methods of the old interface to those of the new. Many functions do not have a name change in the new interface, and are not listed here. Not all functions are available for all cluster types.
| Previous Name | New Name |
|---|---|
findResource | parcluster |
createJob | createJob (no
change) |
createParallelJob | createCommunicatingJob(where 'Type' = 'SPMD') |
createMatlabPoolJob | createCommunicatingJob(where 'Type' = 'Pool') |
destroy | delete |
clearLocalPassword | logout |
createTask | createTask (no
change) |
getAllOutputArguments | fetchOutputs |
getJobSchedulerData | getJobClusterData |
setJobSchedulerData | setJobClusterData |
getCurrentJobmanager | getCurrentCluster |
getFileDependencyDir | getAttachedFilesFolder |
Properties
In addition to a few new properties on the objects in the new interface, some other properties have new names when using the new interface, according to the following tables.
| Previous Scheduler Properties | New Cluster Properties |
|---|---|
DataLocation | JobStorageLocation |
Configuration | Profile |
HostAddress | AllHostAddresses |
Computer | ComputerType |
ClusterOsType | OperatingSystem |
IsUsingSecureCommunication | HasSecureCommunication |
SchedulerHostname, MasterName, Hostname | Host |
ParallelSubmissionWrapperScript | CommunicatingJobWrapper |
ClusterSize | NumWorkers |
NumberOfBusyWorkers | NumBusyWorkers |
NumberOfIdleWorkers | NumIdleWorkers |
SubmitFcn | IndependentSubmitFcn |
ParallelSubmitFcn | CommunicatingSubmitFcn |
DestroyJobFcn | DeleteJobFcn |
DestroyTaskFcn | DeleteTaskFcn |
| Previous Job Properties | New Job Properties |
|---|---|
FileDependencies | AttachedFiles |
PathDependencies | AdditionalPaths |
MinimumNumberOfWorkers, MaximumNumberOfWorkers | NumWorkersRange |
| Previous Task Properties | New Task Properties |
|---|---|
MaximumNumberOfRetries | MaximumRetries |
Getting Help
In addition to these changes, there are some new properties and methods, while some old properties are not used in the new interface. For a list of the methods and properties available for clusters, jobs, and tasks, use the following commands for help on each class type:
help parallel.Cluster help parallel.Job help parallel.Task help parallel.job.CJSIndependentJob help parallel.job.CJSCommunicatingJob help parallel.task.CJSTask help parallel.job.MJSIndependentJob help parallel.job.MJSCommunicatingJob help parallel.task.MJSTask
There might be slight changes in the supported format for properties whose names are still the same. To get help on an individual method or property, the general form of the command is:
help parallel.obj-type.method-or-property-name
You might need to specify the subclass in some cases, for example, where different cluster types use properties differently. The following examples display the help for specified methods and properties of various object types:
help parallel.cluster.LSF.JobStorageLocation help parallel.Job.fetchOutputs help parallel.job.MJSIndependentJob.FinishedFcn help parallel.Task.delete
Compatibility Considerations
Jobs
This release still supports the old form of interface,
however, the old interface and the new interface are not compatible
in the same job. For example, a job manager scheduler object that
you create with findResource requires that you
use only the old interface methods and properties, and cannot be used
for creating a communicating job (createCommunicatingJob).
A cluster that was defined with parcluster must
use the new interface, and cannot be used to create a parallel job
(createParallelJob).
Graphical interfaces provide access only to the new interface. The Configurations Manager is no longer available and is replaced by the Cluster Profile Manager. Actions that use profiles automatically convert and upgrade your configurations to profiles. Therefore, if you already have a group of configurations, the first time you open the Cluster Profile Manager, it should already be populated with your converted profiles. Furthermore, you can specify cluster profiles when using the old interface in places where configurations are expected.
One job manager (or MJS) can accommodate jobs of both the old and new interface at the same time.
Creating and Finding Jobs and Tasks
In the old interface, to create or find jobs or tasks, you could
specify property name and value pairs in structures or cell arrays,
and then provide the structure or cell array as an input argument
to the function you used. In the new interface, property names and
values must be pairs of separate arguments, with the property name
as a string expression and its value of the appropriate type. This
applies to the functions createJob, createCommunicatingJob, createTask, findJob,
and findTask.
Batch
Jobs created by the batch command use the
new interface, even if you specify configurations or properties using
the old interface. For example, the following code generates two identical
jobs of the new interface, even though job j2 is
defined with an old interface property. The script randScript contains
just the one line of code, R = rand(3), and the
default profile is local.
j1 = batch('randScript','AdditionalPaths','c:\temp');
j1.wait;
R1 = j1.load('R');or
j2 = batch('randScript','PathDependencies','c:\temp');
j2.wait;
R2 = j2.load('R');
whos
Name Size Bytes Class
R1 1x1 248 struct
R2 1x1 248 struct
j1 1x1 112 parallel.job.CJSIndependentJob
j2 1x1 112 parallel.job.CJSIndependentJob
Communicating Job Wrapper
In the old interface, for a parallel job in an LSF, TORQUE,
or PBS Pro scheduler, you would call the scheduler's setupForParallelExecution method
with the necessary arguments so that the toolbox could automatically
set the object's ClusterOSType and ParallelSubmissionWrapperScript properties,
thus determining which wrapper was used. In the new interface, with
a communicating job you only have to set the LSF, Torque, or PBSPro
cluster object's OperatingSystem and CommunicatingJobWrapper properties,
from which the toolbox calculates which wrapper to use. For more information
about these properties and their possible values, in MATLAB type
help parallel.cluster.LSF.CommunicatingJobWrapper
You can change the LSF to PBSPro or Torque in
this command, as appropriate.
Enhanced Example Scripts
An updated set of example scripts is available in the product for using a generic scheduler with the new programming interface. These currently supported scripts are provided in the folder:
matlabroot/toolbox/distcomp/examples/integration
For more information on these scripts and their updates, see
the README file provided in each subfolder, or
see Supplied
Submit and Decode Functions.
Compatibility Considerations
Scripts that use the old programming interface are provided
in the folder matlabroot/toolbox/distcomp/examples/integration/old
Cluster Profiles
New Cluster Profile Manager
Cluster profiles replace parallel configurations for defining settings for clusters and their jobs. You can access profiles and the Cluster Profile Manager from the desktop Parallel menu. From this menu you can select or import profiles. To choose a profile as the default, select the desktop menu Parallel > Select Cluster Profile. The current default profile is indicated with a bold dot.

The Cluster Profile Manager lets you create, edit, validate, import, and export profiles, among other actions. To open the Cluster Profile Manager, select Parallel > Manage Cluster Profiles.

For more information about cluster profiles and the Cluster Profile Manager, see Cluster Profiles.
Programming with Profiles
These commands provide access to profiles and the ability to create cluster objects.
| Function | Description |
|---|---|
p = | List of your profiles |
parallel.defaultClusterProfile | Specifies which profile to use by default |
c = | Creates cluster object, c, for accessing
parallel compute resources |
c. | Saves changes of cluster property values to its current profile |
c. | Saves cluster properties to a profile of the specified name, and sets that as the current profile for this cluster |
parallel.importProfile | Import profiles from specified .settings file |
parallel.exportProfile | Export profiles to specified .settings file |
Profiles in Compiled Applications
Because compiled applications include the current profiles of
the user who compiles, in most cases the application has the profiles
it needs. When other profiles are needed, a compiled application can
also import profiles that have been previously exported to a .settings file.
The new ParallelProfile key supports exported parallel
configuration .mat files and exported cluster profile .settings files;
but this might change in a future release. For more information, see Export
Profiles for MATLAB Compiler.
Compatibility Considerations
In past releases, when a compiled application imported a parallel configuration, that configuration would overwrite a configuration of the same if it existed. In this release, imported profiles are renamed if profiles already exist with the same name; so the same profile might have a different name in the compiled application than it does in the exported profile file.
Enhanced GPU Support
GPUArray Support
The following functions are added to those that support GPUArray data, or are enhanced in their support for this release:
beta betaln bsxfun circshift det eig fft fft2 fftn fprintf full | ifft ifft2 ifftn ind2sub int2str inv ipermute isequaln issorted mat2str max | mldivide min mrdivide norm num2str permute qr shiftdim sprintf sub2ind |
Note the following enhancements and restrictions to some of these functions:
GPUArray usage now supports all data types supported by MATLAB, except
int64anduint64.For the list of functions that
bsxfunsupports, see thebsxfunreference page.The full range of syntax is now supported for
fft,fft2,fftn,ifft,ifft2, andifftn.eignow supports all matrices, symmetric or not.issortedsupports only vectors, not matrices.maxandmincan now return two output arguments, including an index vector.mldividesupports complex arrays. It also supports overdetermined matrices (with more rows than columns), with no constraints on the second input.mrdividesupports underdetermined matrices (more columns than rows) as the second input argument.normnow supports the formnorm(X,2), whereXis a matrix.
The following functions are not methods of the GPUArray class, but they do work with GPUArray data:
angle fliplr flipud flipdim fftshift | ifftshift kron mean perms | rank squeeze rot90 trace |
Reset or Deselect GPU Device
Resetting a GPU device clears your GPUArray and CUDAKernel data
from the device. There are two ways to reset a GPU device, while still
keeping it as the currently selected device. You can use the reset function,
or you can use gpuDevice(idx)idx. For
example, use reset:
idx = 1 g = gpuDevice(idx) reset(g)
Alternatively, call gpuDevice again with
the same index argument:
idx = 1 g = gpuDevice(idx) . . . g = gpuDevice(idx) % Resets GPU device, clears data
To deselect the current device, use gpuDevice([ ]) with
an empty argument (as opposed to no argument). This clears the GPU
of all arrays and kernels, and invalidates variables in the workspace
that point to such data.
Asynchronous GPU Calculations and Wait
All GPU calculations now run asynchronously with MATLAB.
That is, when you initiate a calculation on the GPU, MATLAB continues
execution while the GPU runs its calculations at the same time. The wait command
now accommodates a GPU device, so that you can synchronize MATLAB and
the GPU. The form of the command is
wait(gpudev)
where gpudev is the object representing the
GPU device to wait for. At this command, MATLAB waits until all
current calculations complete on the specified device.
Compatibility Considerations
In previous releases, MATLAB and the GPU were synchronous,
so that any calls to the GPU had to complete before MATLAB proceeded
to the next command. This is no longer the case. Now MATLAB continues
while the GPU is running. The wait command lets
you time GPU code execution.
Verify GPUArray or CUDAKernel Exists on the Device
The new function existsOnGPU lets
you verify that a GPUArray or CUDAKernel exists on the current GPU
device, and that its data is accessible from MATLAB. It is possible
to reset the GPU device, so that a GPUArray or CUDAKernel object variable
still exists in your MATLAB workspace even though it is no longer
available on the GPU. For example, you can reset a GPU device using
the command gpuDevice(index)reset(dev)
index = 1;
g = gpuDevice(index);
R = parallel.gpu.GPUArray.rand(4,4)
0.5465 0.3000 0.4067 0.6110
0.9024 0.8965 0.6635 0.7709
0.8632 0.7481 0.9901 0.0420
0.2307 0.7008 0.7516 0.5059
existsOnGPU(R)
1
reset(g); % Resets GPU device
existsOnGPU(R)
0
R % View GPUArray contents
Data no longer exists on the GPU.Any attempt to use the data from R generates
an error.
MATLAB Code on the GPU
GPU support is extended to include the following MATLAB code
in functions called by arrayfun to
run on the GPU:
and beta betaln eq ge gt int8 int16 intmax | intmin ldivide le lt minus ne not or plus | power rdivide realmax realmin times uint8 uint16 |
Note the following enhancements and restrictions to some of these functions:
plus,minus,ldivide,rdivide,power,times, and other arithmetic, comparison or logical operator functions were previously supported only when called with their operator symbol. Now they are supported in their functional form, so can be used as direct argument inputs toarrayfunandbsxfun.arrayfunandbsxfunon the GPU now support the integer data typesint8,uint8,int16, anduint16.
The code in your function can now call any functions defined in your function file or on the search path. You are no longer restricted to calling only those supported functions listed in the table of Supported MATLAB Code.
Set CUDA Kernel Constant Memory
The new setConstantMemory method on the
CUDAKernel object lets you set kernel constant memory from MATLAB.
For more information, see the setConstantMemory reference
page.
Latest NVIDIA CUDA Device Driver
This version of Parallel Computing Toolbox GPU functionality supports only the latest NVIDIA CUDA device driver.
Compatibility Considerations
Earlier versions of the toolbox supported earlier versions of CUDA device driver. Always make sure you have the latest CUDA device driver.
Enhanced Distributed Array Support
Newly Supported Functions
The following functions now support distributed arrays with all forms of codistributor (1-D and 2DBC), or are enhanced in their support for this release:
Random Number Generation on Workers
MATLAB worker sessions now generate random number values using the combined multiplicative recursive generator (mrg32k3a) by default.
Compatibility Considerations
In past releases, MATLAB workers used the same default generator as a MATLAB client session. This is no longer the case.
New Job Monitor
The Job Monitor is a tool that lets you track the jobs you have submitted to a cluster. It displays the jobs for the scheduler determined by your selection of a parallel configuration. Open the Job Monitor from the MATLAB desktop by selecting Parallel > Job Monitor.
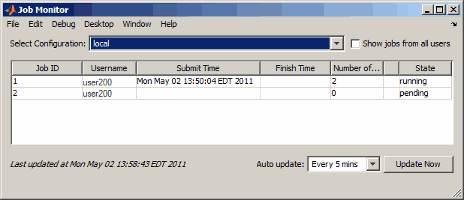
Right-click a job in the list to select a command from the context menu for that job:
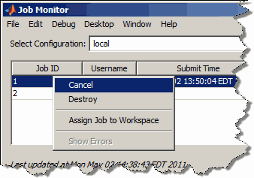
For more information about the Job Monitor and its capabilities, see Job Monitor.
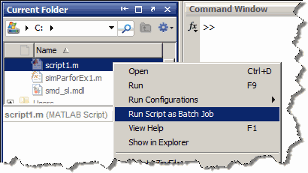
Run Scripts as Batch Jobs from the Current Folder Browser
From the Current Folder browser, you can run a MATLAB script
as a batch job by browsing to the file's folder, right-clicking
the file, and selecting Run Script as Batch Job.
The batch job runs on the cluster identified by the current default
parallel configuration. The following figure shows the menu option
to run the script from the file script1.m:
Number of Local Workers Increased to Twelve
You can now run up to 12 local workers on your MATLAB client
machine. If you do not specify the number of local workers in a command
or configuration, the default number of local workers is determined
by the value of the local scheduler's ClusterSize property,
which by default equals the number of computational cores on the client
machine.
Enhanced GPU Support
Latest NVIDIA CUDA Device Driver
This version of Parallel Computing Toolbox GPU functionality supports only the latest NVIDIA CUDA device driver.
Compatibility Considerations
Earlier versions of the toolbox supported earlier versions of CUDA device driver. Always make sure you have the latest CUDA device driver.
Deployment of GPU Applications
MATLAB Compiler™ generated standalone executables and components now support applications that use the GPU.
Random Number Generation
You can now directly create arrays of random numbers on the GPU using these new static methods for GPUArray objects:
parallel.gpu.GPUArray.rand parallel.gpu.GPUArray.randi parallel.gpu.GPUArray.randn
The following functions set the GPU random number generator seed and stream:
parallel.gpu.rng parallel.gpu.RandStream
Also, arrayfun called with GPUArray data
now supports rand, randi, and randn.
For more information about using arrayfun to generate
random matrices on the GPU, see Generating
Random Numbers on the GPU.
GPUArray Support
The following functions now support GPUArray data:
chol diff eig find isfinite isinf | isnan lu max min mldivide | norm not repmat sort svd |
mldivide supports complex arrays. It also
supports overdetermined matrices (with more rows than columns) when
the second input argument is a column vector (has only one column).
eig supports only symmetric matrices.
max and min return only
one output argument; they do not return an index vector.
The following functions are not methods of the GPUArray class, but they do work with GPUArray data:
angle beta betaln fliplr flipud | flipdim fftshift ifftshift kron | mean perms squeeze rot90 |
The default display of GPUArray variables now shows the array contents. In previous releases, the display showed some of the object properties, but not the contents. For example, the new enhanced display looks like this:
M = gpuArray(magic(3))
M =
8 1 6
3 5 7
4 9 2
To see that M is a GPUArray, use the whos or class function.
MATLAB Code on the GPU
GPU support is extended to include the following MATLAB code
in functions called by arrayfun to
run on the GPU:
Also, the handle passed to arrayfun can reference
a simple function, a subfunction, a nested function, or an anonymous
function. The function passed to arrayfun can call
any number of its subfunctions. The only restriction is that when
running on the GPU, nested and anonymous functions do not have access
to variables in the parent function workspace. For more information
on function structure and relationships, see Types of Functions.
Enhanced Distributed Array Support
Newly Supported Functions
The following functions are enhanced to support distributed arrays, supporting all forms of codistributor (1-D and 2DBC):
The following functions can now directly construct codistributed arrays:
codistributed.linspace(m, n, ..., codist)codistributed.logspace(m, n, ..., codist)
Conversion of Error and Warning Message Identifiers
For R2011b, error and warning message identifiers have changed in Parallel Computing Toolbox.
Compatibility Considerations
If you have scripts or functions that use message identifiers
that changed, you must update the code to use the new identifiers.
Typically, message identifiers are used to turn off specific warning
messages, or in code that uses a try/catch statement
and performs an action based on a specific error identifier.
For example, the 'distcomp:old:ID' identifier
has changed to 'parallel:similar:ID'. If your code
checks for 'distcomp:old:ID', you must update it
to check for 'parallel:similar:ID' instead.
To determine the identifier for a warning, run the following command just after you see the warning:
[MSG,MSGID] = lastwarn;
This command saves the message identifier to the variable MSGID.
To determine the identifier for an error, run the following commands just after you see the error:
exception = MException.last; MSGID = exception.identifier;
Tip Warning messages indicate a potential issue with your code. While you can turn off a warning, a suggested alternative is to change your code so that it runs warning-free. |
Task Error Properties Updated
If there is an error during task execution, the task Error property
now contains the non-empty MException object that
is thrown. If there was no error during task execution, the Error property
is empty.
The identifier and message properties of this object are now
the same as the task's ErrorIdentifier and ErrorMessage properties,
respectively. For more information about these properties, see the Error, ErrorIdentifier,
and ErrorMessage reference
pages.
Compatibility Considerations
In past releases, when there was no error, the Error property
contained an MException object with empty data fields, generated by MException('',
''). Now to determine if a task is error-free, you can query
the Error property itself to see if it is empty:
didTaskError = ~isempty(t.Error)
where t is the task object you are examining.
Deployment of Local Workers
MATLAB Compiler generated standalone executables and libraries from parallel applications can now launch up to eight local workers without requiring MATLAB Distributed Computing Server software.
New Desktop Indicator for MATLAB Pool Status
When you first open a MATLAB pool from your desktop session, an indicator appears in the lower-right corner of the desktop to show that this desktop session is connected to an open pool. The number indicates how many workers are in the pool.
![]()
When you close the pool, the indicator remains displayed and shows a value of 0.
Enhanced GPU Support
Static Methods to Create GPUArray
The following new static methods directly create GPUArray objects:
parallel.gpu.GPUArray.linspace parallel.gpu.GPUArray.logspace
GPUArray Support
The following functions are enhanced to support GPUArray data:
cat colon conv conv2 cumsum cumprod eps filter filter2 horzcat meshgrid ndgrid plot subsasgn subsindex subsref vertcat
and all the plotting related functions.
GPUArray Indexing
Because GPUArray now supports subsasgn and subsref,
you can index into a GPUArray for assigning and reading individual
elements.
For example, create a GPUArray and assign the value of an element:
n = 1000; D = parallel.gpu.GPUArray.eye(n); D(1,n) = pi
Create a GPUArray and read the value of an element back into the MATLAB workspace:
m = 500; D = parallel.gpu.GPUArray.eye(m); one = gather(D(m,m))
MATLAB Code on the GPU
GPU support is extended to include the following MATLAB code
in functions called by arrayfun to run on the GPU:
&, |, ~, &&, ||, while, if, else, elseif, for, return, break, continue, eps
You can now call eps with string inputs,
so your MATLAB code running on the GPU can include eps('single') and eps('double').
NVIDIA CUDA Driver 3.2 Support
This version of Parallel Computing Toolbox GPU functionality supports only NVIDIA CUDA device driver 3.2.
Compatibility Considerations
Earlier versions of the toolbox supported earlier versions of CUDA device driver. If you have an older driver, you must upgrade to CUDA device driver version 3.2.
Distributed Array Support
Newly Supported Functions
The following functions are enhanced to support distributed arrays, supporting all forms of codistributor (1-D and 2DBC):
arrayfun cat reshape
Enhanced mtimes Support
The mtimes function
now supports distributed arrays that use a 2-D block-cyclic (2DBC)
distribution scheme, and distributed arrays that use 1-D distribution
with a distribution dimension greater than 2. Previously, mtimes supported
only 1-D distribution with a distribution dimension of 1 or 2.
The mtimes function now returns a distributed
array when only one of its inputs is distributed, similar to its behavior
for two distributed inputs.
Compatibility Considerations
In previous releases, mtimes returned a replicated
array when one input was distributed and the other input was replicated.
Now it returns a distributed array.
Enhanced parfor Support
Nested for-Loops Inside parfor
You can now create nested for-loops inside
a parfor-loop, and you can use both the parfor-loop
and for-loop variables directly as indices for
the sliced array inside the nested loop. See Nested
Loops.
Enhanced Support for Microsoft Windows HPC Server
Support for 32-Bit Clients
The parallel computing products now support Microsoft Windows HPC Server on 32-bit Windows clients.
Search for Cluster Head Nodes Using Active Directory
The findResource function can search Active
Directory to identify your cluster head node. For more information,
see the findResource reference
page.
Enhanced Admin Center Support
You can now start and stop mdce services on remote hosts from Admin Center. For more information, see Start mdce Service.
New Remote Cluster Access Object
New functionality is available that lets you mirror job data
from a remote cluster to a local data location. This supports the
generic scheduler interface when making remote submissions to a scheduler
or when using a nonshared file system. For more information, see the RemoteClusterAccess object
reference page.
GPU Computing
This release provides the ability to perform calculations on a graphics processing unit (GPU). Features include the ability to:
Use a GPU array interface with several MATLAB built-in functions so that they automatically execute with single- or double-precision on the GPU — functions including
mldivide,mtimes,fft, etc.Create kernels from your MATLAB function files for execution on a GPU
Create kernels from your CU and PTX files for execution on a GPU
Transfer data to/from a GPU and represent it in MATLAB with GPUArray objects
Identify and select which one of multiple GPUs to use for code execution
For more information on all of these capabilities and the requirements to use these features, see GPU Computing.
Job Manager Security and Secure Communications
You now have a choice of four security levels when using the job manager as your scheduler. These levels range from no security to user authentication requiring passwords to access jobs on the scheduler.
You also have a choice to use secure communications between the job manager and workers.
For more detailed descriptions of these features and information about setting up job manager security, see Set MJS Cluster Security.
The default setup uses no security, to match the behavior of past releases.
Generic Scheduler Interface Enhancements
Decode Functions Provided with Product
Generic scheduler interface decode functions for distributed and parallel jobs are now provided with the product. The two decode functions are named:
parallel.cluster.generic.distributedDecodeFcn parallel.cluster.generic.parallelDecodeFcn
These functions are included on the workers' path. If your submit
functions make use of the definitions in these decode functions, you
do not have to provide your own decode functions. For example, to
use the standard decode function for distributed jobs, in your submit
function set MDCE_DECODE_FUNCTION to 'parallel.cluster.generic.distributedDecodeFcn'.
For information on using the generic scheduler interface with submit
and decode functions, see Use the Generic
Scheduler Interface.
Enhanced Example Scripts
This release provides new sets of example scripts for using the generic scheduler interface. As in previous releases, the currently supported scripts are provided in the folder
matlabroot/toolbox/distcomp/examples/integration
In this location there is a folder for each type of scheduler:
lsf— Platform LSF®pbs— PBSsge— Sun™ Grid Enginessh— generic UNIX-based scriptswinmpiexec— mpiexec on Windows
For the updated scheduler folders (lsf, pbs, sge),
subfolders within each specify scripts for different cluster configurations: shared, nonshared, remoteSubmission.
For more information on the scripts and their updates, see the README file
provided in each folder, or see Supplied
Submit and Decode Functions.
Compatibility Considerations
For those schedulers types with updated scripts in this release
(lsf, pbs, sge),
the old versions of the scripts are provided in the folder matlabroot/toolbox/distcomp/examples/integration/old
batch Now Able to Run Functions
Batch jobs can now run functions as well as scripts. For more
information, see the batch reference
page.
batch and matlabpool Accept Scheduler Object
The batch function and the functional form
of matlabpool now accept a scheduler object as
their first input argument to specify which scheduler to use for allocation
of compute resources. For more information, see the batch and matlabpool reference
pages.
Enhanced Functions for Distributed Arrays
qr Supports Distributed Arrays
The qr function
now supports distributed arrays. For restrictions on this functionality,
type
help distributed/qr
mldivide Enhancements
The mldivide function
(\) now supports rectangular distributed arrays.
Formerly, only square matrices were supported as distributed arrays.
When operating on a square distributed array, if the second
input argument (or right-hand side of the operator) is replicated, mldivide now
returns a distributed array.
Compatibility Considerations
In previous releases, mldivide returned a
replicated array when the second (or right-hand side) input was replicated.
Now it returns a distributed array.
chol Supports 'lower' option
The chol function
now supports the 'lower' option when operating
on distributed arrays. For information on using chol with
distributed arrays, type
help distributed/chol
eig and svd Return Distributed Array
When returning only one output matrix, the eig and svd functions
now return a distributed array when the input is distributed. This
behavior is now consistent with outputs when requesting more than
one matrix, which returned distributed arrays in previous releases.
Compatibility Considerations
In previous releases, eig and svd returned
a replicated array when you requested a single output. Now they return
a distributed array if the output is a single matrix. The behavior
when requesting more than one output is not changed.
transpose and ctranspose Support 2dbc
In addition to their original support for 1-D distribution schemes,
the functionsctranspose and transpose now
support 2-D block-cyclic ('2dbc') distributed arrays.
Inf and NaN Support Multiple Formats
Distributed and codistributed arrays now support nan, NaN, inf and Inf for
not-a-number and infinity values with the following functions:
| Infinity Value | Not-a-Number |
|---|---|
codistributed.Inf | codistributed.NaN |
codistributed.inf | codistributed.nan |
distributed.Inf | distributed.NaN |
distributed.inf | distributed.nan |
Support for Microsoft Windows HPC Server 2008 R2
Parallel Computing Toolbox software now supports Microsoft Windows HPC Server 2008 R2. There is no change in interface compared to using HPC Server 2008. Configurations and other toolbox utilities use the same settings to support both HPC Server 2008 and HPC Server 2008 R2.
User Permissions for MDCEUSER on Microsoft Windows
The user identified by the MDCEUSER parameter
in the mdce_def file is now granted all necessary
privileges on a Windows system when you install the mdce process.
For information about what permissions are granted and how to reset
them, see Set
the User.
Compatibility Considerations
In past releases, you were required to set the MDCEUSER permissions
manually. Now this is done automatically when installing the mdce
process.
Compatibility Summary
| Release | Features or Changes with Compatibility Considerations |
|---|---|
| R2015b | |
| R2015a | |
| R2014b | |
| R2014a | |
| R2013b |
|
| R2013a | None |
| R2012b | gpuArray Class Name |
| R2012a | |
| R2011b | |
| R2011a | |
| R2010b |