Table of Contents
Here is a description of the coordinates sytems used in Aims and Anatomist, and what I have understood of how SPM handles its referentials.
Anatomist uses AIMS to handle its referentials so behaves exactly the same way.
Aims tries to work internally in an image-specific referential, but with always the same orientation. This orientation is axial with the following coordinates system:
- X axis: right to left
- Y axis: front to back
- Z axis: top to bottom
- origin: the center of the first voxel: the voxel in the top, right, front corner
Once loaded in memory, all voxels should be organized in this order. As a consequence, images in Anatomist are always displayed in radiological mode, whatever the actual orientation of data on disk.
Images on disk, depending on their format and acquisition modes, are not necessarily in this orientation. When a different orientation is detected, images are flipped in memory at load-time to fit the standard AIMS orientation. And when images are written back to disk, they may also be flipped back according to the specific format needs.
By default, AIMS doesn't apply any transformation other than flipping images at load time as described just before.
But transformations can be provided in some Aims commands or loaded in Anatomist to apply coordinates changes. Then coords transformations are applied on the fly when processing or displaying data which are not in the same referential.
There is no special referential (such as a common central working referential).
Transformation files used by AIMS (.trm files) are ASCII files looking like this:
Tx Ty Tz R11 R12 R13 R21 R22 R23 R31 R32 R33Tx, Ty, Tz are the translation components while the Rij coefficients are the linear matrix part. When used, these coefficients are applied as a "standard" 4x4 transformation matrix:
[ R11 R12 R13 Tx ]
M = [ R21 R22 R23 Ty ]
[ R31 R32 R33 Tz ]
[ 0 0 0 1 ]
AIMS (and Anatomist) writes an additional header file which can store any additional information: the .minf header (for Meta-INFormation) when saving its data (images, meshes, and any other data), and reads it if it is present when loading data files. This meta-header has the shape displayed by the AimsFileInfo command, and may be saved in "python dictionary" or XML formats. The MINF file has the same file name as the main data file, with the .minf extension added (toto.img.minf for instance).
The MINF header may contain referentials and transformations information. When present, this information is stored in a few fields:
referentialmay store an unique identifier (a cryptic characters string) to identify the AIMS referential for the current data file. If several data files refer to the same identifier, then they share the same referential and are considered to have coordinates in the same system.referentialsmay store a list of target referentials for transformations specified in thetransformationsfield. Both fields must have the same number of entries. Referentials are identified by character strings, either as unique identifiers or generic names (not necessarily unique). Some standard common referentials have specific names: "Talairach-MNI template-SPM" for the MNI normalization referential (used by SPM for instance), or "Talairach-AC/PC-Anatomist" for the referential based on anterior and posterior commissures used by the BrainVISA anatomical segmentation pipeline.transformationsmay store a list of transformation matrices, each going from the AIMS data referential to the corresponding referential specified in thereferentialsfield (same position in the list). Each transformation is a 4x4 matrix written as 16 numbers in rows, and assumes all coordinates are in millimeters.storage_to_memorymay store the disk orientation information, by providing the transformation between the disk storage voxels order and the memory orientation (the AIMS referential). This transformation is in the same shape as thetransformationsfield, except that it is not a list, and the transformation is in voxels, not in mm.
attributes = {
'storage_to_memory' : [ 1, 0, 0, 0, 0, -1, 0, 62, 0, 0, -1, 45, 0, 0, 0, 1 ],
'referentials' : [ 'Coordinates aligned to another file or to anatomical truth' ],
'transformations' : [ [ -1, 0, 0, 78, 0, -1, 0, 75, 0, 0, -1, 84, 0, 0, 0, 1 ] ],
'referential': 'be9724cc-eceb-d831-a83e-335e12b80f14',
}
The referentials and transformations information in the MINF header may reflect information already stored in the specific format header (Analyze origin, or NIFTI-1 qform and sform, or MINC transformation).
Internally, SPM thinks things are always in the same orientation, which is also axial but with different axes:
- X axis: left to right
- Y axis: back to front
- Z axis: bottom to top
- origin: the center of the voxel specified by the origin field of the SPM image header. This origin is specified in voxels and starts counting from 1 (not 0) like a matlab array index does.
Working on the coordinate transformations for years and regularly getting headaches from it, I am still not 100% sure of what I say here, so if I'm wrong, please correct me by sending a message on BrainVisa forum (http://brainvisa.info/forum/). Especially, I'm not sure that SPM99 and SPM2 really use the same referentials.
SPM handles input Analyze images in two different orientations: axial radiological and axial neurological orientations. This orientation is not specified in SPM-Analyze format image files, so you have to tell how they are oriented. This is done in SPM by a flipping flag set somewhere in SPM defaults configuration (default.analyze.flip in SPM2).
This is specific to SPM-Analyze format, and does not apply to NIFTI-1 or Minc formats. Hopefully the Analyze format is now obsolete and will disapear with time, but there are still existing files...
This flipping flag has changed in form and meaning between SPM99 and SPM2.
As I have understood:
- SPM99:
- SPM99 uses the flipping flag only when normalizing images, indicating that the normalization process must perform or not a flip towards the normalization template. A clear indication of it is that the flag is part of the normalization parameters and is not present in other parts of SPM.
- Otherwise, SPM99 does not bother about the orientation of images. This is to say: even when displaying images, radiological images will be displayed with the left on the right of the display window, and neurological images with the left on the left, regardless of the flipping flag.
- Normalized images are always in neurological orientation whatever the orientation of input unnormalized images. Consequently, after normalizing a radiological image, loading both a normalized image and an unnormalized one in SPM will display them with different orientations.
- I am not sure if normalization templates have to be necessarily in neurological orientation or not but I guess yes because there is no way to indicate that the template is in radiological orientation.
- Normalization matrices for radiological data contain a X axis flip (negative 1st coefficient)
- SPM2:
- SPM2 uses the flipping flag at load time: radiological images are systematically flipped when loaded (and flipped back when rewritten so as to keep their radiological orientation on disk). This is true for all Analyze/SPM image files.
- This means all processings use it. As a consequence, all images are displayed in neurological orientation, left on the left, even for radiological images.
- But as this flag is global in SPM, all SPM images are considered to be in the same orientation: you cannot mix radio and neuro images. What I am pointing out here is only valid for SPM format images: SPM2 also handles MINC format, and Minc images contain orientation information.
- Normalized images are now in the same orientation as the input unnormalized image. Normalizing a radiological image will result in a normalized file in the radiological orientation (in SPM format). This is not what SPM99 used to do.
- SPM2 does not understand SPM99 and vice versa: no compatibility at all (neither forward nor backward): if you are using the radiological convention (like we are), loading in SPM2 an image normalized by SPM99 will result in a spurious flip and incorrect display and processing. This means you cannot use with SPM2 an image database built with SPM99.
- Normalization templates can be in either orientation. More precisely, I guess the template must be in the orientation specified by the flipping flag, or in Minc format in neurological orientation. I'm not completely sure of this. But this is perhaps an explanation of why the standard normalization template is now in Minc format and not in SPM format (otherwise its interpretation would depend on a user-defined flag).
Last minute: I have just discovered that SPM now sometimes produces images with negative voxel sizes. I guess it is a kind of flipping indication, but I don't know from what to what else. And we know that all radiological images don't have this negative voxel size feature. So my opinion is that it's not reliable at all (at least unless you exactly know which version of SPM has written each image and this info is not available). This sign information is ignored in the current version of AIMS.
The headache goes on...
- SPM5 ans SPM8:
- SPM5 now uses the NIFTI-1 format for all output. NIFTI-1 specifies orientations and possibly transformations to standard referentials in its format, so this is a very good thing. Many problems are now solved.
- Otherwise I guess SPM5 behaves essentially like SPM2.
- The only little imperfection is that when SPM5 performs normalizations towards the MNI template, it does not indicate in output image that the target referential is the MNI template, but an unspecified other referential instead. So you have to know the target referential and specify it when needed (for instance in Anatomist).
SPM uses a common central referential to work in. Every image can provide a transformation matrix to this referential. Such a transformation may be specified in different ways:
- an optional
.matfile with the same name as the SPM format image. this was the way SPM99 and SPM2 behaved with Analyze format. But with SPM5 and SPM8, using NIFTI-1 formats avoids this need. - If this
.matis not provided, then the file format header information is considered. NIFTI-1 provides full affine transformation matrices, but Analyze has only the origin translation, which is considered to be the only transformation needed to reach the central referential. If the .mat file is specified, information contained in it overrides some of the header information (including the origin).
Normalization files (*_sn3d.mat for SPM99, *_sn.mat for SPM2 and newer) contain transformations to the referential of a normalization template (either a standard one provided with the SPM software distribution, or a custom user-made one). This transformation contains an affine part (matrix), and optionally, depending on the normalization type, a non-linear part (coefficients on a functions base as far as I know but I don't know much about this part). Information about the input and template images are also included (dimensions, and origins or voxels-to-template transformation).
Normalized images are in the referential of the normalization template used, but not necessarily with the same bounding box, resolution and field of view.
SPM99 and SPM2 use normalization files with different names and different contents. They are not compatible, even if there is some common and similar information in them.
Due to the different internal orientations of the coordinate systems, going from SPM to AIMS and vice versa causes some serious problems.
SPM normalization files are in matlab (.mat) format. AIMS cannot read the proprietary matlab format, so such files cannot be directly imported in AIMS.
However, the scipy module for Python language can read them. So we have made Python scripts in PyAims and in BrainVisa to convert SPM matrices to AIMS .trm format. Only the affine part can be converted, because AIMS only use matrices for transformations, and non-linear information cannot fit into a matrix. Look at the AimsSpmNormalizationConvert.py program, and the SPMsn3dToAims process in BrainVISA.
As the orientation is different in SPM and AIMS, a transformation to a template image is not the same as a transformation to a normalized image with a different field of view. So, when converting SPM normalization matrices, the normalized image must be also provided, otherwise BrainVisa can only give the transformation to the normalization template. Note the difference.
It is impossible to guess the orientation of such images if you don't know how they were acquired. This means you have to manually specify their orientation, either for all images in SPM, or in BrainVisa when importing them into a database. BrainVisa tags them so it knows everything afterwards and avoids mistakes. SPM does not.
Normalizing the same image in radiological orientation with SPM99 and SPM2 results in normalized images in different orientations. When you import normalized images coming from another site, you have to know whether they have been normalized by SPM99 or by SPM2, and if the original image was in radiological or neurological orientation.
I think the normalization file (*_sn3d.mat for SPM99, or *_sn.mat for SPM2) contains enough information to retreive the orientation of input and template images, so can disambiguate the situation.
The origin field of SPM format is the position of the referential origin, in voxels and starting from 1 (not from 0). In fact it's a matlab array index. So it is given in the orientation of the image on disk. AIMS flips SPM images on several axes when loading them, so the origin information also has to be flipped. Flipping it needs to know the image dimensions.
AIMS referentials have their origin in the first voxel, (almost) in the corner of the image, and normally don't use the SPM origin. But the origin information is read and maintained. Anatomist can, if asked for, make a transformation going from AIMS origin (corner) to the SPM origin. This allows to display several aligned SPM images in Anatomist with the correct correspondance. However after this translation, the coordinates are still in AIMS orientation (radiological and indirect), not in SPM, so the coordinates do not correspond to what they are in SPM.
To compare coordinates of SPM images in Anatomist and SPM, another transformation has to be applied in Anatomist, with all the flips included. Anatomist can directly use the SPM/MNI normalization referential.
Up to now, GIS images are considered being always in AIMS orientation unless specified in their AIMS meta-header (.minf file, see the corresponding paragraph). No flips are applied.
The Minc and NIFTI-1 IO plugins take orientation into account and flip data accordingly when reading / writing files.
I am not sure if other formats (Dicom, Ecat...) can specify an image orientation or not. If they do, the current release of AIMS will probably not take it into account.
SPM99 and SPM2 don't use the same format of normalization files, but both provide more or less the following information:
-
An affine transformation matrix, called
Affine, transforming coordinates from the template space to the input space, both sides in voxels arrays index, and indexed from 1 (not from 0) -
A voxels-to-mm transformation matrix for the input image, transforming voxels of the image into a mm position in the SPM internal orientation, taking the origin into account, and possibly rotations if the format supports it (NIFTI). This matrix is called
VF.matin SPM2 and also performs flipping, and calledMFin SPM99 but doesn't seem to contain the flipping information. However for nomrmalization this millimetric referential is quite undefined and we will not really use it. -
Another voxel-to-mm matrix for the template image:
VG.matin SPM2, orMGin SPM99. -
Input and template image dimensions in voxels and a bit more:
VF.dimandVG.dimin SPM2, orDimsin SPM99.
- 3 images: input (I call it Anatomy to be clearer), template, and normalized images. I use the suffixes A, T and N for coordinates on these 3 images.
-
I use the same name for a given referential and coordinates in this referential: for instance
RAAis both the AIMS referential of the anatomical image and a coordinates vector in it. I don't bother about standardized math notations: I don't remember them and haven't been using math anymore for many years. Don't ask me too much. -
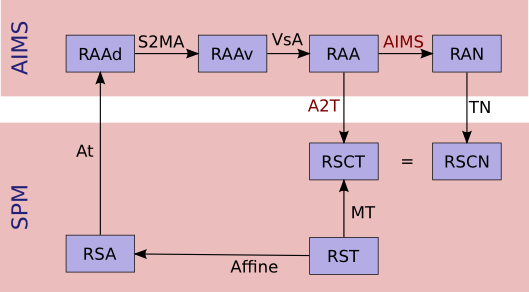
AIMS referentials:
RAA: anatomy (in mm, radio convention, origin in 1st voxel)RAAv: anatomy (in voxels, radio convention, origin in 1st voxel)RAAd: anatomy (in voxels, disk storage ordering)RAN: normalized (in mm...)
-
SPM referentials:
RSA: anatomy, in voxelsRST: template, in voxelsRSCT: template, "central" in mmRSCN: normalized, "central" in mm. Actually,RSCTandRSCNare the same.
- Transformation matrices:
Affine: the SPM affine matrix (voxels):RSTtoRSAAt: SPM voxels to AIMS voxels transformation. This is only to take the array indexing starting at 1 in Matlab. SoAtis a( -1, -1, -1 )voxel translation. It can be used betweenRSAandRAAd, and either betweenRSNandRANd.AIMS:RAAtoRAN, what we want to calculate. Here again, I'm maybe not using correctly math notations. I mean:RAN = AIMS * RAA.VsA: Aims voxels to mm anatS2MAAims "storage_to_memory" anat matrix: disk voxels to Aims voxels.A2T: Aims anat-mm to template space-mm:RAAtoRSCT, what we want to calculate if no normalized image is used.TN: normalized, Aims-mm to SPM-central-mm:RANtoRSCNTCN: template to normalized in SPM-mm:RSCTtoRSCN. This transformation is identity in fact because the template and normalized images are in the same referential internally in SPM, but it's maybe clearer if I mention it.