Introduction to Parallel Solutions
Interactively Run a Loop in Parallel
This section shows how to modify a simple for-loop
so that it runs in parallel. This loop does not have a lot of iterations,
and it does not take long to execute, but you can apply the principles
to larger loops. For these simple examples, you might not notice an
increase in execution speed.
Suppose your code includes a loop to create a sine wave and plot the waveform:
for i = 1:1024 A(i) = sin(i*2*pi/1024); end plot(A)
You can modify your code to run your loop in parallel by using a
parforstatement:parfor i = 1:1024 A(i) = sin(i*2*pi/1024); end plot(A)
The only difference in this loop is the keyword
parforinstead offor. When the loop begins, it opens a parallel pool of MATLAB® sessions called workers for executing the iterations in parallel. After the loop runs, the results look the same as those generated from the previousfor-loop.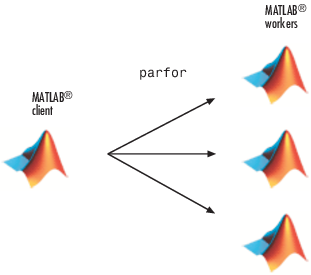
Because the iterations run in parallel in other MATLAB sessions, each iteration must be completely independent of all other iterations. The worker calculating the value for
A(100)might not be the same worker calculatingA(500). There is no guarantee of sequence, soA(900)might be calculated beforeA(400). (The MATLAB Editor can help identify some problems withparforcode that might not contain independent iterations.) The only place where the values of all the elements of the arrayAare available is in your MATLAB client session, after the data returns from the MATLAB workers and the loop completes.
For more information on parfor-loops, see Parallel for-Loops (parfor).
You can modify your cluster profiles to control how many workers run your loops, and whether the workers are local or on a cluster. For more information on profiles, see Clusters and Cluster Profiles.
Modify your parallel preferences to control whether a parallel pool is created automatically, and how long it remains available before timing out. For more information on preferences, see Parallel Preferences.
You can run Simulink® models in parallel loop iterations
with the sim command inside
your loop. For more information and examples of using Simulink with parfor,
see Run Parallel Simulations in the Simulink documentation.
Run a Batch Job
To offload work from your MATLAB session to run in the
background in another session, you can use the batch command. This example uses the for-loop
from the previous example, inside a script.
To create the script, type:
edit mywaveIn the MATLAB Editor, enter the text of the
for-loop:for i = 1:1024 A(i) = sin(i*2*pi/1024); end
Save the file and close the Editor.
Use the
batchcommand in the MATLAB Command Window to run your script on a separate MATLAB worker:job = batch('mywave')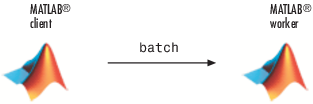
The
batchcommand does not block MATLAB, so you must wait for the job to finish before you can retrieve and view its results:wait(job)
The
loadcommand transfers variables created on the worker to the client workspace, where you can view the results:load(job,'A') plot(A)When the job is complete, permanently delete its data and remove its reference from the workspace:
delete(job) clear job
batch runs your code on a local worker
or a cluster worker, but does not require a parallel pool.
You can use batch to run either scripts
or functions. For more details, see the batch reference
page.
Run a Batch Parallel Loop
You can combine the abilities to offload a job and run a parallel
loop. In the previous two examples, you modified a for-loop
to make a parfor-loop, and you submitted a script
with a for-loop as a batch job. This example combines
the two to create a batch parfor-loop.
Open your script in the MATLAB Editor:
edit mywaveModify the script so that the
forstatement is aparforstatement:parfor i = 1:1024 A(i) = sin(i*2*pi/1024); end
Save the file and close the Editor.
Run the script in MATLAB with the
batchcommand as before, but indicate that the script should use a parallel pool for the loop:job = batch('mywave','Pool',3)
This command specifies that three workers (in addition to the one running the batch script) are to evaluate the loop iterations. Therefore, this example uses a total of four local workers, including the one worker running the batch script. Altogether, there are five MATLAB sessions involved, as shown in the following diagram.
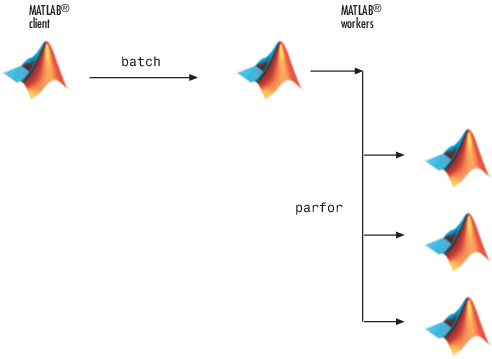
To view the results:
wait(job) load(job,'A') plot(A)The results look the same as before, however, there are two important differences in execution:
The work of defining the
parfor-loop and accumulating its results are offloaded to another MATLAB session bybatch.The loop iterations are distributed from one MATLAB worker to another set of workers running simultaneously (
'Pool'andparfor), so the loop might run faster than having only one worker execute it.
When the job is complete, permanently delete its data and remove its reference from the workspace:
delete(job) clear job
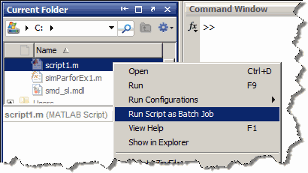
Run Script as Batch Job from the Current Folder Browser
From the Current Folder browser, you can run a MATLAB script
as a batch job by browsing to the file's folder, right-clicking
the file, and selecting Run Script as Batch Job.
The batch job runs on the cluster identified by the default cluster
profile. The following figure shows the menu option to run the script
file script1.m:
Running a script as a batch from the browser uses only one worker
from the cluster. So even if the script contains a parfor loop
or spmd block, it does not open an additional
pool of workers on the cluster. These code blocks execute on the single
worker used for the batch job. If your batch script requires opening
an additional pool of workers, you can run it from the command line,
as described in Run a Batch Parallel Loop.
When you run a batch job from the browser, this also opens the Job Monitor. The Job Monitor is a tool that lets you track your job in the scheduler queue. For more information about the Job Monitor and its capabilities, see Job Monitor.
Distribute Arrays and Run SPMD
Distributed Arrays
The workers in a parallel pool communicate with each other, so you can distribute an array among the workers. Each worker contains part of the array, and all the workers are aware of which portion of the array each worker has.
Use the distributed function to distribute an
array among the workers:
M = magic(4) % a 4-by-4 magic square in the client workspace
MM = distributed(M)Now MM is a distributed array, equivalent
to M, and you can manipulate or access its elements
in the same way as any other array.
M2 = 2*MM; % M2 is also distributed, calculation performed on workers x = M2(1,1) % x on the client is set to first element of M2
Single Program Multiple Data (spmd)
The single program multiple data (spmd) construct
lets you define a block of code that runs in parallel on all the workers
in a parallel pool. The spmd block can run on some
or all the workers in the pool.
spmd % By default creates pool and uses all workers R = rand(4); end
This code creates an individual 4-by-4 matrix, R,
of random numbers on each worker in the pool.
Composites
Following an spmd statement, in the client
context, the values from the block are accessible, even though the
data is actually stored on the workers. On the client, these variables
are called Composite objects. Each element of
a composite is a symbol referencing the value (data) on a worker in
the pool. Note that because a variable might not be defined on every
worker, a Composite might have undefined elements.
Continuing with the example from above, on the client, the Composite R has
one element for each worker:
X = R{3}; % Set X to the value of R from worker 3.The line above retrieves the data from worker 3 to assign the
value of X. The following code sends data to worker
3:
X = X + 2;
R{3} = X; % Send the value of X from the client to worker 3.If the parallel pool remains open between spmd statements
and the same workers are used, the data on each worker persists from
one spmd statement to another.
spmd R = R + labindex % Use values of R from previous spmd. end
A typical use for spmd is to run the same
code on a number of workers, each of which accesses a different set
of data. For example:
spmd INP = load(['somedatafile' num2str(labindex) '.mat']); RES = somefun(INP) end
Then the values of RES on the workers are
accessible from the client as RES{1} from worker
1, RES{2} from worker 2, etc.
There are two forms of indexing a Composite, comparable to indexing a cell array:
AA{n}returns the values ofAAfrom workern.AA(n)returns a cell array of the content ofAAfrom workern.
Although data persists on the workers from one spmd block
to another as long as the parallel pool remains open, data does not
persist from one instance of a parallel pool to another. That is,
if the pool is deleted and a new one created, all data from the first
pool is lost.
For more information about using distributed arrays, spmd,
and Composites, see Distributed Arrays and SPMD.