Requirement: if no database is set, add a database
For more details about data importation, see BrainVISA manual - Data importation chapter
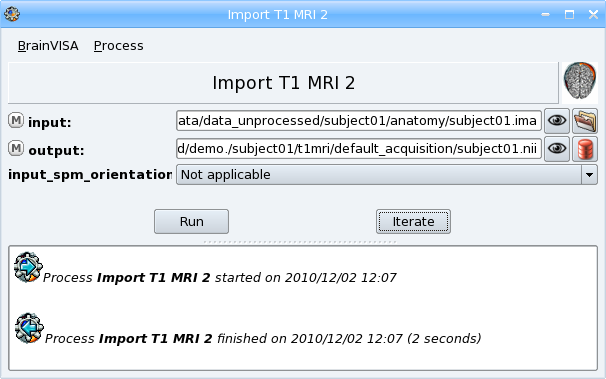
- Open the process Morphologist => import => Import T1 MRI.
- Select input data with
 icon : data_unprocessed/subject01/anatomy/subject01.ima.
icon : data_unprocessed/subject01/anatomy/subject01.ima. - Describe the output with
 icon. You have to fill in several information fields for this data
icon. You have to fill in several information fields for this data
- Select a format: for example NIFTI-1 image; the image will be converted to this format in the database directory.
- Type a name for the protocol attribute: for example demo
- A suggested item appears in the right panel. Click Ok button to accept.
- Click Run button to start the process.
To replay the movie, right-click on the movie to display the Adobe Flash Player menu. Run back then play again the movie.
Note
In this example, the raw data is in GIS format but it is possible to import data which are in various formats including Dicom. To import a Dicom image, select the first file of the serie as input. You may have to change the file filter to All files (*) in the file browser to see the Dicom files if they have not the extension .dcm. Data will be converted to GIS or Nifti format in BrainVISA database. See the paragraph about importation of Dicom images in BrainVISA manual for more details.The image has been copied in the BrainVISA database directory. Now, let us have a look at the database and its content:
- Click on the
 icon to visualize the output image with Anatomist. Click again on the button to close the visualization. Right-click on the button to open the viewer process associated to this type of data.
icon to visualize the output image with Anatomist. Click again on the button to close the visualization. Right-click on the button to open the viewer process associated to this type of data. - Open the process Data management => Database browser to explore the database directory. Double-click on the database directory to see its content, open the subdirectories (demo/subject01/t1mri/default_acquisition) to find the imported image. Click on the image and see information about it in the right panel. Right-click on the image to open the contextual menu. Select view to vizualize the image with Anatomist. More information about the database browser in the BrainVISA manual.
Requirement: if there is no T1 MRI in the database Import a T1 MRI
In this step, we are going to spot 4 points in the image: the anterior commissure, the posterior commissure, an inter-hemispheric point and a point of the left hemisphere. The spotted coordinates will be written in a .APC file which will be used during the Morphologist pipeline to compute a transformation to the Talairach AC/PC referential, which is useful to see several subjects images in a common referential.
Note
This step can be replaced by a normalization using SPM or FSL. An alternative step using a normalization is offered in Morphologist pipeline if SPM or FSL is installed on your system.
If you want to use the SPM normalization, you have to set the SPM paths in BrainVISA configuration. To do so, go to BrainVISA menu -> Preferences -> SPM. An Auto detect button is available to try and find the paths automatically.
This process is included in the Morphologist pipeline as a first step but it is also possible to run it separately.
- Open the process Morphologist => Segmentation Pipeline => components => Prepare subject for Anatomical Pipeline.
- Select input data with
 icon. The output field
Commissures_coordinates is automatically filled thanks to BrainVISA database system.
icon. The output field
Commissures_coordinates is automatically filled thanks to BrainVISA database system. - In order to fill the coordinates ot the AC, PC points, click on
 button. Anatomist starts and a window containing the T1 MRI image is opened. Scroll the slices of the image to find AC point, click on the point, then click on the
button. Anatomist starts and a window containing the T1 MRI image is opened. Scroll the slices of the image to find AC point, click on the point, then click on the  button near Anterior_commissure field to copy the coordinates of the point. Do the same for the other points. Be careful for the left hemisphere point : Anatomist displays images in radiological mode by default, so left and right are inverted.
button near Anterior_commissure field to copy the coordinates of the point. Do the same for the other points. Be careful for the left hemisphere point : Anatomist displays images in radiological mode by default, so left and right are inverted. - The allow_flip_initial_MRI field by default does not allow to modify the image if it is not correctly oriented. You can change it to True if the image is not correctly oriented.
- Click Run button to start the process.
To replay the movie, right-click on the movie to display the Adobe Flash Player menu. Run back then play again the movie.
Requirement: AC/PC spotting or normalization
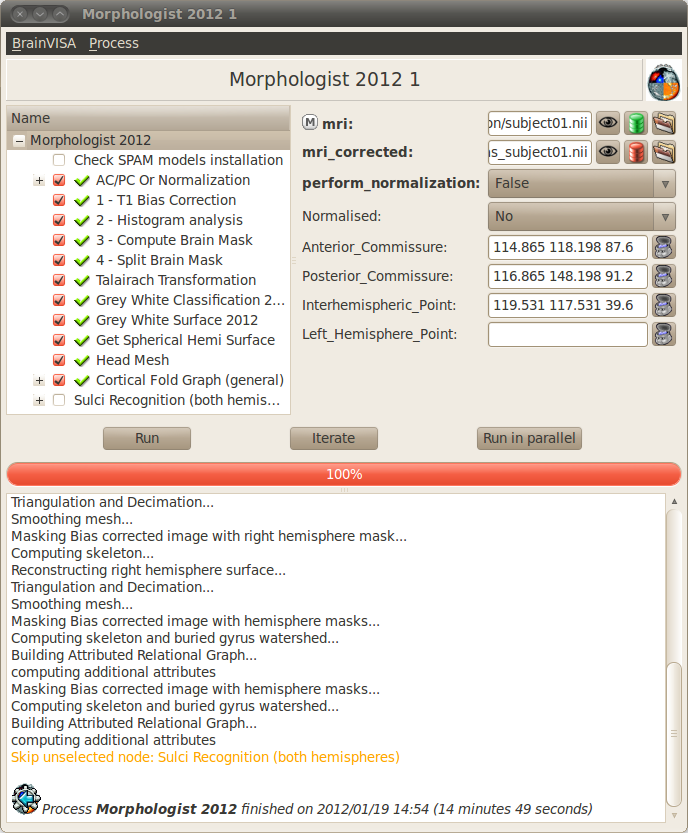
The anatomical pipeline is made of the following processing steps :
T1 Bias correction (mri_corrected) : Corrects for the spatial bias in usual MR images.
Histogram analysis (histo_analysis) : Analyses a T1-weighted histogram to estimate grey/white statistics.
Compute Brain mask (brain_mask) : Computes a binary mask of the brain from a bias corrected T1-weighted image from the histogram analysis.
Split brain mask (split_mask) : Splits the brain into three parts (hemispheres + cerebellum).
Talairach Transformation (Talairach_transform) : Computes a transformation between the image referential and the Talairach AC/PC referential.
Grey White classification: Computes a mask of the grey and white matter (left_grey_white and right_grey_white).
Grey White Surface: Computes a 3D mesh of the grey / white interface for each hemisphere (left_white_mesh and right_white_mesh).
Grey Spherical Hemi Surface: Computes a 3D mesh of the external surface of the cortex for each hemispheres (left_hemi_mesh and right_hemi_mesh).
Head mesh (head_mesh): Creation of the head mesh. Mainly useful for visualization purpose.
Cortical fold graph (left_graph and right_graph): Builds a graph representation of the cortical folds for each hemisphere.
Sulci recognition: automatic labelling of the sulci. It is not selected by default because it can take a long time.
For more information about the pipeline steps, see the slides of a presentation about the Morphologist pipeline (http://brainvisa.info/doc/morphologist/morphologist.pdf).
To run the pipeline :
- Open the process Morphologist => Morphologist 2013. This version of the pipeline is new since version 4.2.0, and is regularly updated in later versions. The older pipelines can still be found in Segmentation pipeline => Older pipelines.
- Select the input T1 MRI with the
 icon.
icon. - All the other parameters are automatically filled in.
- Unselect the first step AC / PC or normalization because it is already done.
- Click Run button to start the process.
It can be useful to keep a trace of the processes applied to data. To do so, you can save a process with its parameters and options in a file:
- In the process window, select Process => Save menu.
- Choose a file name and location (eg. t1_pipeline_subject01.bvproc) and click on the Save button.
- The saved process can be loaded through BrainVISA => Open process menu.
Once a step is finished, its results are written and can be visualized.
- Select a step of the pipeline, its parameters appear in the right panel.
- Click on the
 button near each output data (indicated by the
button near each output data (indicated by the  icon) to visualize it.
icon) to visualize it. - Click again on the
 button to close the visualization.
button to close the visualization.
The new Morphologist pipeline has been greatly improved and should succeed on most images. In case it fails, here are a few advices :
First, let us find which step has failed:
- Look at the error message and find the name of the step which has failed.
- Select the steps before the one that failed and visualize their output parameters to see if there are abnormal results.
To know what happened, you can also read BrainVISA log : click on BrainVISA => Show log menu. A window showing the historic of processes started in the current session appears.
If the automatic segmentation is not perfect, it is possible to correct it manually.
- Select the brain mask segmentation step for subject03
- Click on the
 button near the brain_mask parameter.
button near the brain_mask parameter. - A popup window indicating Click here when finished appears. You will have to click here when the correction is done. For the moment, reduce it.
- The brain mask is loaded in Anatomist as an editable region of interest named label_255. It is shown in a 3D window on the T1 MRI image.
- In the Anatomist toolbox window, change the brush in the Paint tab.
- Add voxels to the mask by maintaining left click and moving the mouse.
- Remove voxels from the mask by pressing Ctrl key while drawing.
- Fix the mask by removing some voxels outside the brain.
- When the correction is over, click on the Ok button of the dialog window indicating Click here when finished.
- The modified mask is saved.
Warning
After a manual correction, do not start again the brain mask segmentation step, else the corrected mask will be erased. To be sure to not erase by mistake a data, it is possible to lock using the contextual menu lock. More information about this locking feature in the BrainVISA manual.Requirement: Morphologist pipeline to obtain sulci graph.
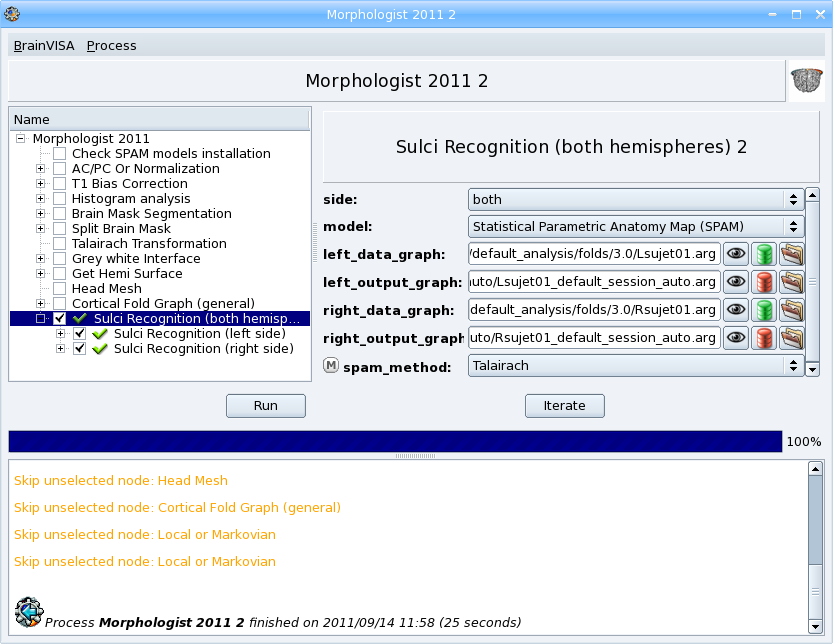
It is available as the last step of the Morphologist pipeline but it can also be run as a separate process. The process is in Morphologist => Sulci => Recognition => Sulci Recognition (both hemispheres).
- In the Morphologist pipeline window, once it is finished, check the box near the Sulci recognition step.
- Right-click to open the contextual menu and choose Unselect before to unselect all the previous steps that are already done.
- In the parameters panel of Sulci recognition process, select Statistical Parametric Anatomy Map (SPAM) for the model.
- Select Talairach for the spam_method parameter.
- Click Run button to start the process.
- When the process is finished, visualize the results.
Warning
The SPAM recognition requires the installation of the SPAM models which are not included in the main BrainVISA package because of their size. These models are in additional packages downloadable on Brainvisa website. They can be easily installed thanks to the process Morphologist -> Sulci -> Recognition -> SPAM models installation. Anyway, a process that checks this installation is now automatically selected as a first of the Morphologist pipeline when the sulci recognition is selected. If the models are not found, Brainvisa will suggest you to install them.
Requirement: Automatic recognition.
This process does not actually make statistics, it only gives some descriptors on regions, according to a model. This process has been primarily written for cortical folds graphs and it is still its main usage, but it can also be used to obtain descriptors for gyri or ROIs.
The input sulci graphs must be labelled. The outputs will be one CSV file for each sulcus with one line per subject.
- Open the process Morphometry => Morphometry statistics.
- Select the input data_graphs with
 button : choose a labelled cortical folds graph of the subject we have just processed (subject01).
button : choose a labelled cortical folds graph of the subject we have just processed (subject01).
- The model is automatically filled in, it depends on the type of graph chosen. For example, the model is not the same for the left and right hemisphere because they does not have the same sulci.
- Choose the output_directory with
 : a directory where the results files will be written. For example, create a new directory named sulci_stats in your personal directory.
: a directory where the results files will be written. For example, create a new directory named sulci_stats in your personal directory. - Click Run button to start the process.
- Once the process is finished, go to the output directoy and open one of the results file with a text editor. The file can also be opened with a spreadsheet software (eg. Excel) to compute statistics.
Note
It is generally more useful to run this process on several subjects to compare their sulci descriptors. We will do that in a next part: Sulci descriptors comparison.In order to have information about the descriptors computed in this process, you can run the viewer of the model. It displays a documentation about the model and the associated descriptors.
To do so:
- Click on the
 button near the model parameter.
button near the model parameter. - If you can see the button, go to the preferences panel and change to user level to Expert. This viewer is in expert user level, so when the level is set to basic, you cannot see it.
- Read the documentation to find the name of the descriptor for the depth of the sulci.
Requirement: Anatomical pipeline to obtain sulci graph.
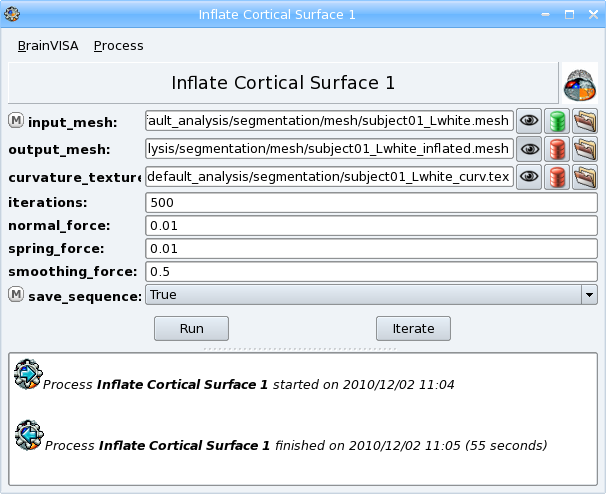
This process inflates the cortical surface for visualization purpose. It can be used for example to visualize an activation map on the cortical surface with Anatomist. Only the white mesh (grey/white interface) can be used as input of this process, the hemisphere mesh (external surface of the cortex) cannot be used because it has not a spherical topology that would enable to apply a deformation.
- Open the process Morphologist => surface => Inflate cortical surface.
- Select an Hemisphere White mesh of subject01 as input_mesh with
 button.
button. - Select True for the save_sequence parameter. The intermediate steps of deformation will be saved in a 4D mesh.
- Click Run button to start the process.
The process has two outputs: the inflated mesh (output_mesh) and a texture containing information about the curvature of the surface.
Once the process is finished, visualize the results by clicking on the  button near the output_mesh parameter. The viewer opens the inflated mesh with the curvature texture applied on it. Move the slider to scroll through the deformation steps of the mesh.
button near the output_mesh parameter. The viewer opens the inflated mesh with the curvature texture applied on it. Move the slider to scroll through the deformation steps of the mesh.