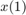lsqcurvefit
Solve nonlinear curve-fitting (data-fitting) problems in least-squares sense
Nonlinear least-squares solver
Find coefficients x that solve the problem
given input data xdata, and the observed output ydata, where xdata and ydata are matrices or vectors, and F (x, xdata) is a matrix-valued or vector-valued function of the same size as ydata.
Optionally, the components of x can have lower and upper bounds lb, and ub. The arguments x, lb, and ub can be vectors or matrices; see Matrix Arguments.
The lsqcurvefit function uses the same
algorithm as lsqnonlin. lsqcurvefit simply
provides a convenient interface for data-fitting problems.
Rather than compute the sum of squares, lsqcurvefit requires
the user-defined function to compute the vector-valued
function
Syntax
- example
x = lsqcurvefit(fun,x0,xdata,ydata) - example
x = lsqcurvefit(fun,x0,xdata,ydata,lb,ub) - example
x = lsqcurvefit(fun,x0,xdata,ydata,lb,ub,options) x = lsqcurvefit(problem)[x,resnorm] = lsqcurvefit(___)- example
[x,resnorm,residual,exitflag,output] = lsqcurvefit(___) [x,resnorm,residual,exitflag,output,lambda,jacobian] = lsqcurvefit(___)
Description
x = lsqcurvefit(fun,x0,xdata,ydata)x0 and finds coefficients x to
best fit the nonlinear function fun(x,xdata) to
the data ydata (in the least-squares sense). ydata must
be the same size as the vector (or matrix) F returned
by fun.
Note:
Passing Extra Parameters explains
how to pass extra parameters to the vector function |
x = lsqcurvefit(fun,x0,xdata,ydata,lb,ub)x,
so that the solution is always in the range lb ≤ x ≤ ub.
You can fix the solution component x(i) by specifying lb(i) = ub(i).
Note:
If the specified input bounds for a problem are inconsistent,
the output Components of |
x = lsqcurvefit(problem)problem, where problem is
a structure described in Input Arguments.
Create the problem structure by exporting a problem
from Optimization app, as described in Exporting Your Work.
Input Arguments
Output Arguments
Limitations
The Levenberg-Marquardt algorithm does not handle bound constraints.
The trust-region-reflective algorithm does not solve underdetermined systems; it requires that the number of equations, i.e., the row dimension of F, be at least as great as the number of variables. In the underdetermined case,
lsqcurvefituses the Levenberg-Marquardt algorithm.Since the trust-region-reflective algorithm does not handle underdetermined systems and the Levenberg-Marquardt does not handle bound constraints, problems that have both of these characteristics cannot be solved by
lsqcurvefit.lsqcurvefitcan solve complex-valued problems directly with thelevenberg-marquardtalgorithm. However, this algorithm does not accept bound constraints. For a complex problem with bound constraints, split the variables into real and imaginary parts, and use thetrust-region-reflectivealgorithm. See Fit a Model to Complex-Valued Data.The preconditioner computation used in the preconditioned conjugate gradient part of the trust-region-reflective method forms JTJ (where J is the Jacobian matrix) before computing the preconditioner. Therefore, a row of J with many nonzeros, which results in a nearly dense product JTJ, can lead to a costly solution process for large problems.
If components of x have no upper (or lower) bounds,
lsqcurvefitprefers that the corresponding components ofub(orlb) be set toinf(or-inffor lower bounds) as opposed to an arbitrary but very large positive (or negative for lower bounds) number.
You can use the trust-region reflective algorithm in lsqnonlin, lsqcurvefit,
and fsolve with small- to medium-scale
problems without computing the Jacobian in fun or
providing the Jacobian sparsity pattern. (This also applies to using fmincon or fminunc without
computing the Hessian or supplying the Hessian sparsity pattern.)
How small is small- to medium-scale? No absolute answer is available,
as it depends on the amount of virtual memory in your computer system
configuration.
Suppose your problem has m equations and n unknowns.
If the command J = sparse(ones(m,n)) causes
an Out of memory error on your machine,
then this is certainly too large a problem. If it does not result
in an error, the problem might still be too large. You can find out
only by running it and seeing if MATLAB runs within the amount
of virtual memory available on your system.
More About
References
[1] Coleman, T.F. and Y. Li. "An Interior, Trust Region Approach for Nonlinear Minimization Subject to Bounds." SIAM Journal on Optimization, Vol. 6, 1996, pp. 418–445.
[2] Coleman, T.F. and Y. Li. "On the Convergence of Reflective Newton Methods for Large-Scale Nonlinear Minimization Subject to Bounds." Mathematical Programming, Vol. 67, Number 2, 1994, pp. 189–224.
[3] Dennis, J. E. Jr. "Nonlinear Least-Squares." State of the Art in Numerical Analysis, ed. D. Jacobs, Academic Press, pp. 269–312.
[4] Levenberg, K. "A Method for the Solution of Certain Problems in Least-Squares." Quarterly Applied Mathematics 2, 1944, pp. 164–168.
[5] Marquardt, D. "An Algorithm for Least-squares Estimation of Nonlinear Parameters." SIAM Journal Applied Mathematics, Vol. 11, 1963, pp. 431–441.
[6] Moré, J. J. "The Levenberg-Marquardt Algorithm: Implementation and Theory." Numerical Analysis, ed. G. A. Watson, Lecture Notes in Mathematics 630, Springer Verlag, 1977, pp. 105–116.
[7] Moré, J. J., B. S. Garbow, and K. E. Hillstrom. User Guide for MINPACK 1. Argonne National Laboratory, Rept. ANL–80–74, 1980.
[8] Powell, M. J. D. "A Fortran Subroutine for Solving Systems of Nonlinear Algebraic Equations." Numerical Methods for Nonlinear Algebraic Equations, P. Rabinowitz, ed., Ch.7, 1970.